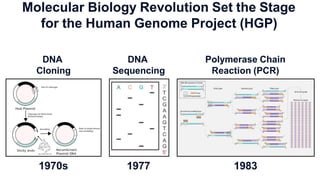
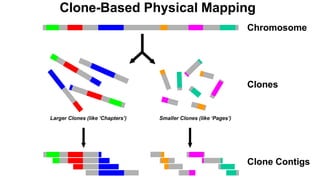
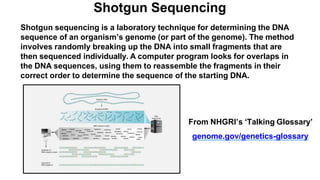
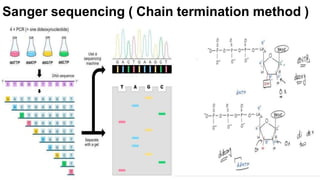
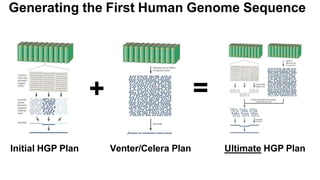
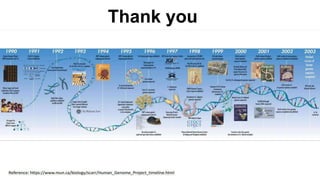
The document outlines the Human Genome Project (HGP), initiated in the late 1980s, with the goal of sequencing the human genome and identifying all human genes. It highlights key contributions from various institutions, challenges faced during sequencing, and the methods used, including Sanger and shotgun sequencing. The HGP was completed ahead of schedule in 2003, resulting in a high-quality sequence of over 90% of the human genome, paving the way for future genomic research.



![1986
1984
Drumbeat of Discussions Leading Up to HGP
1987
1988 1988 1989
“For the newly developing discipline
of [genome] mapping/sequencing
(including the analysis of the
information), we have adopted the
term GENOMICS…
Genomics](https://image.slidesharecdn.com/humangenomeproject-231123095316-02da54ea/85/HUMAN-GENOME-PROJECT-pptx-4-320.jpg)