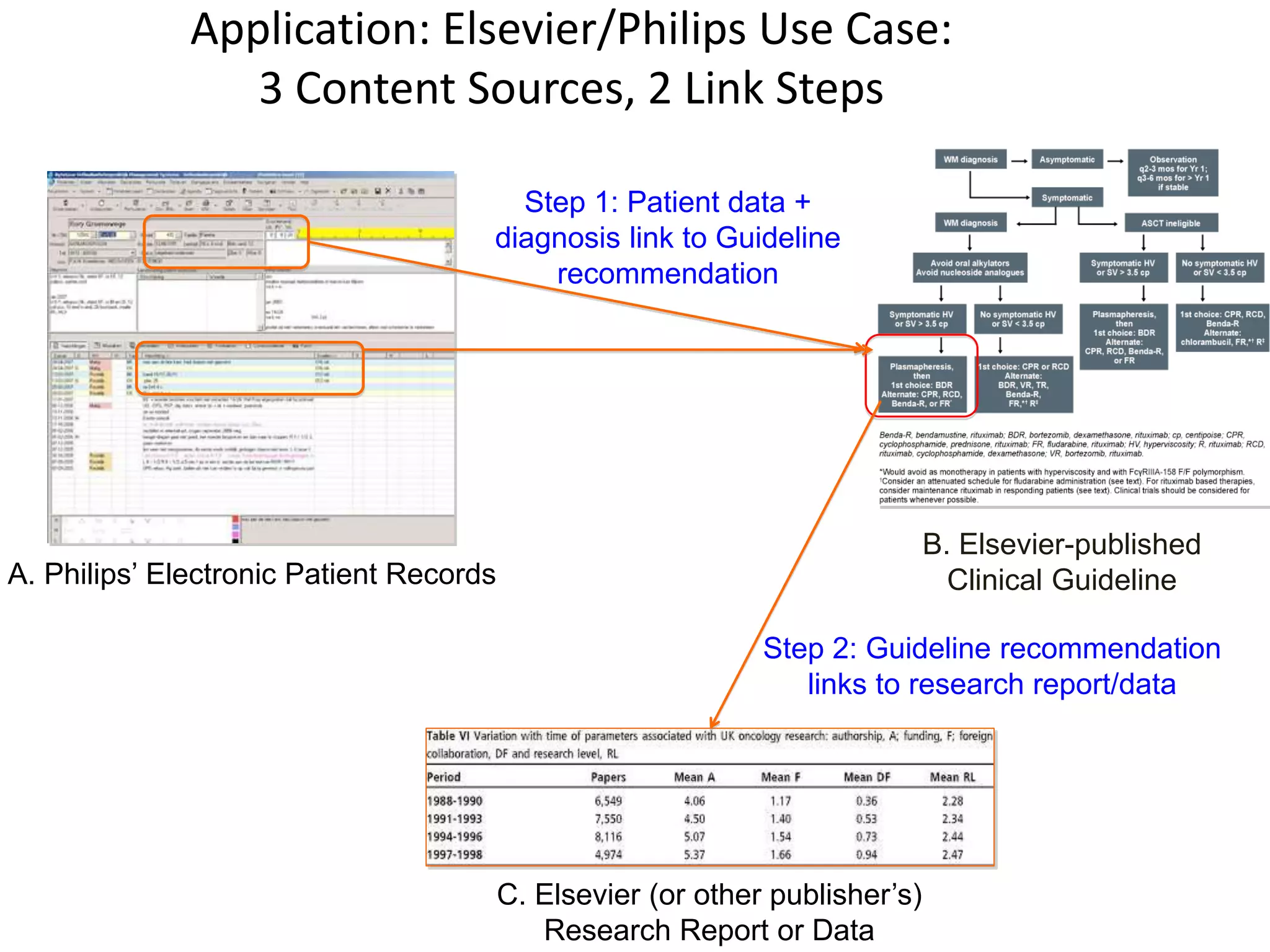
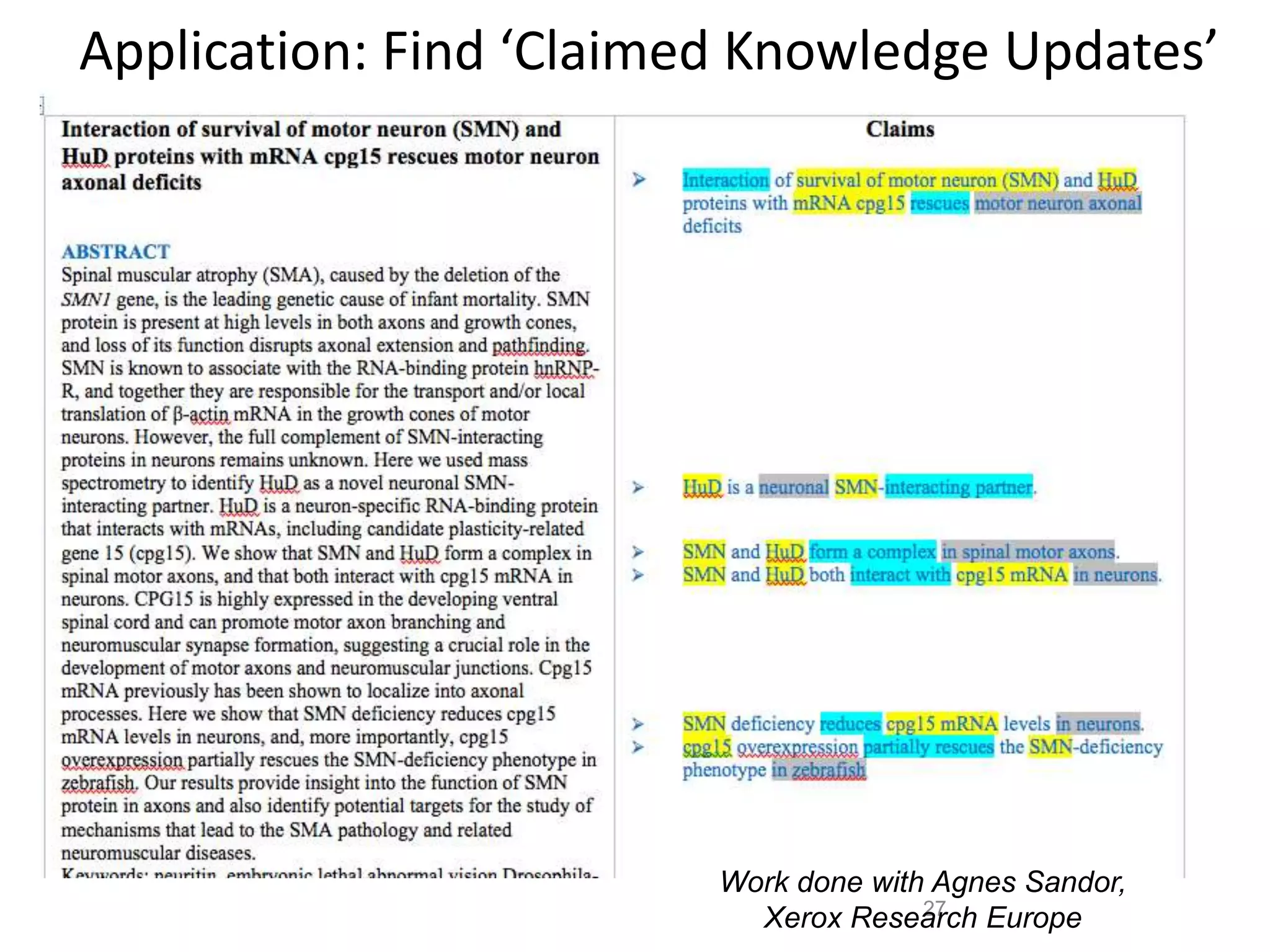
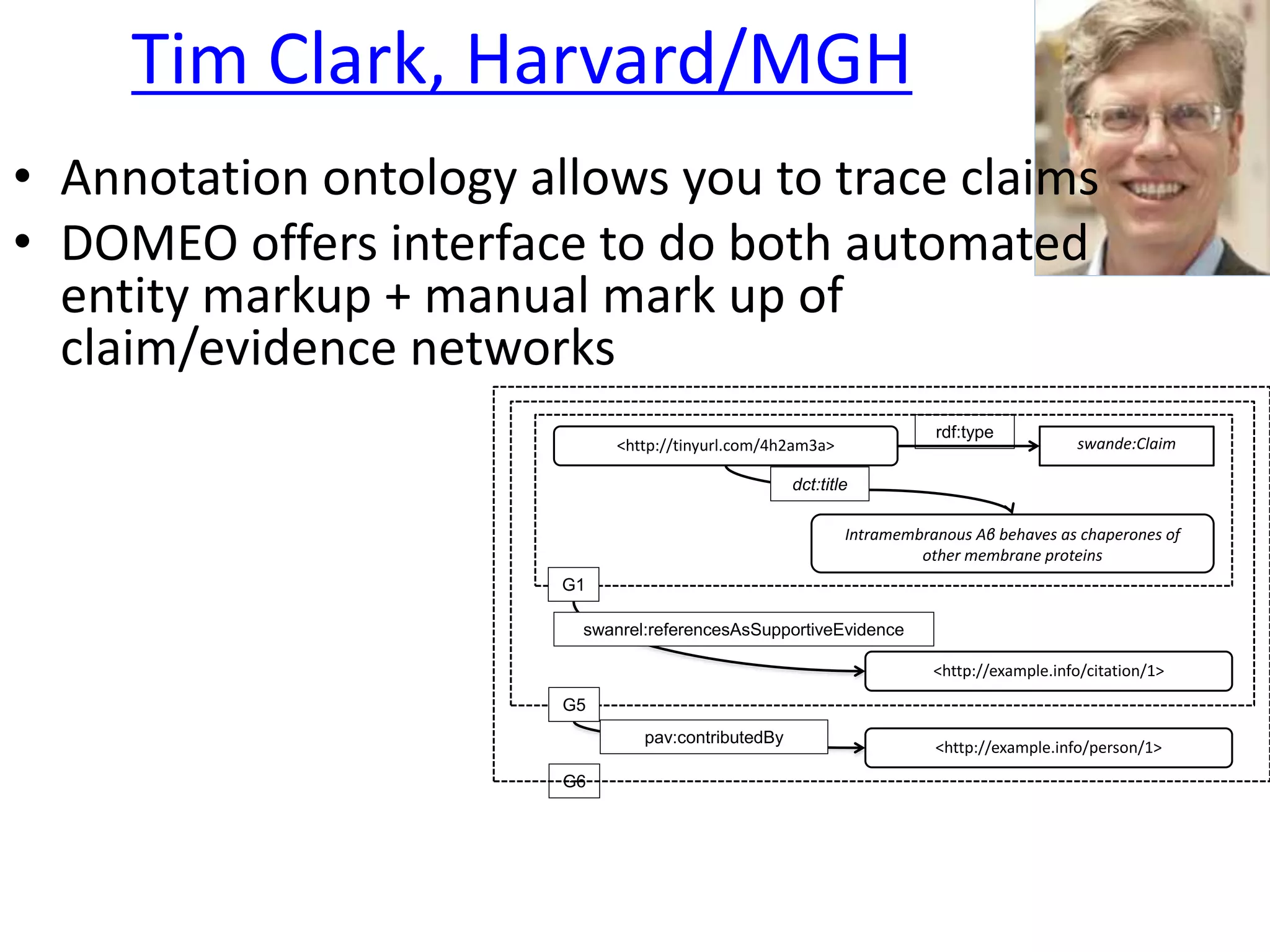
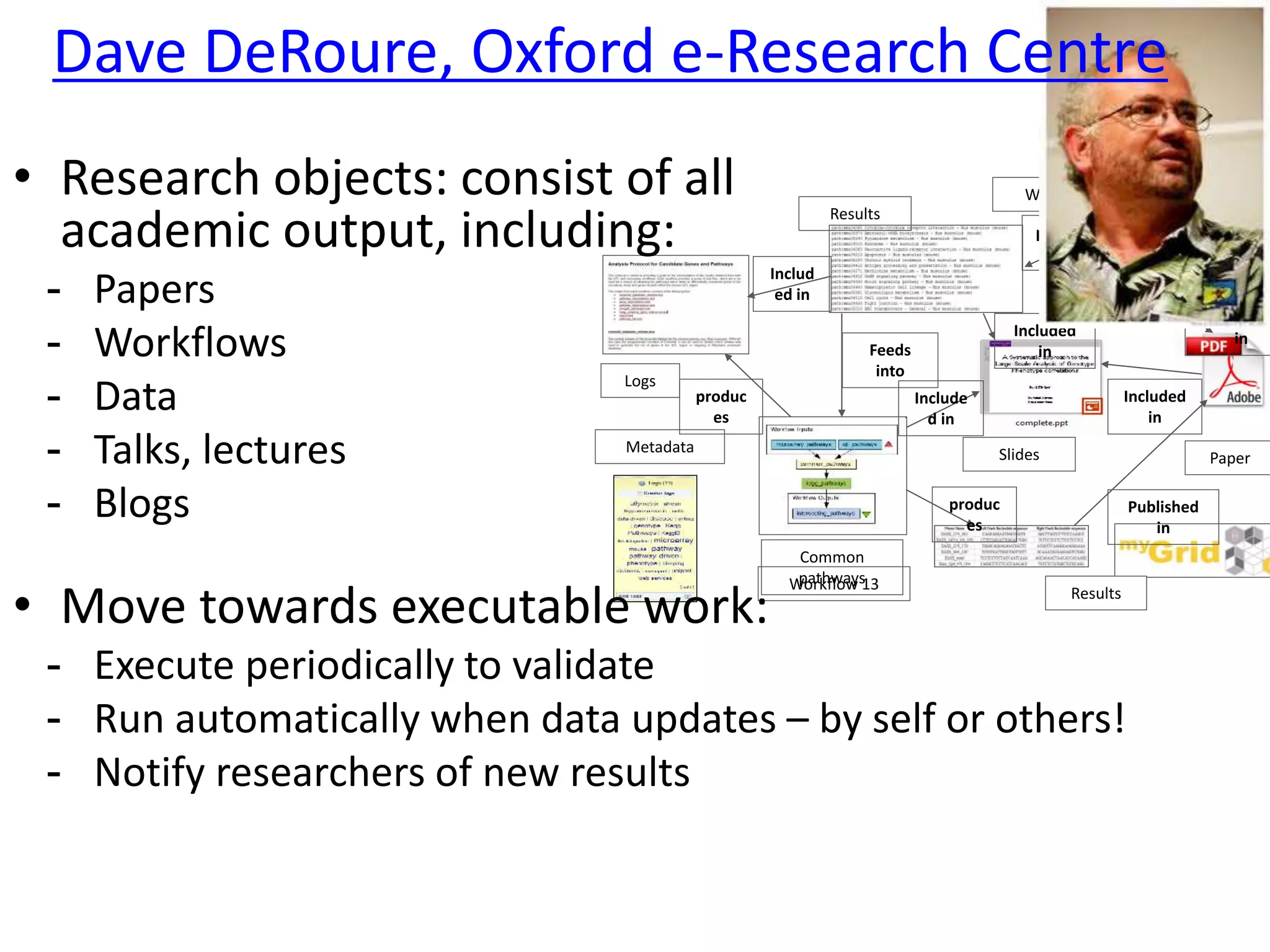
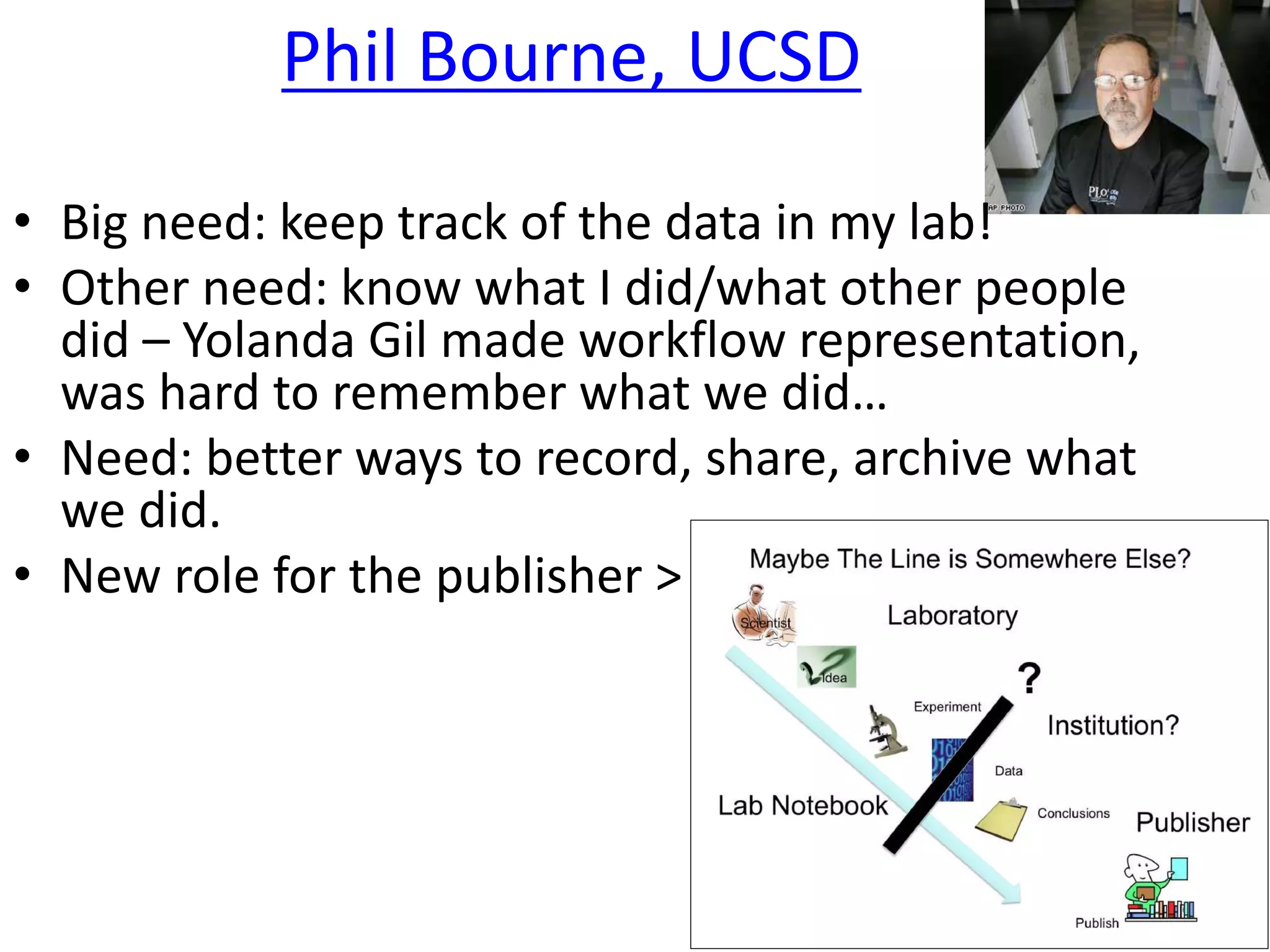
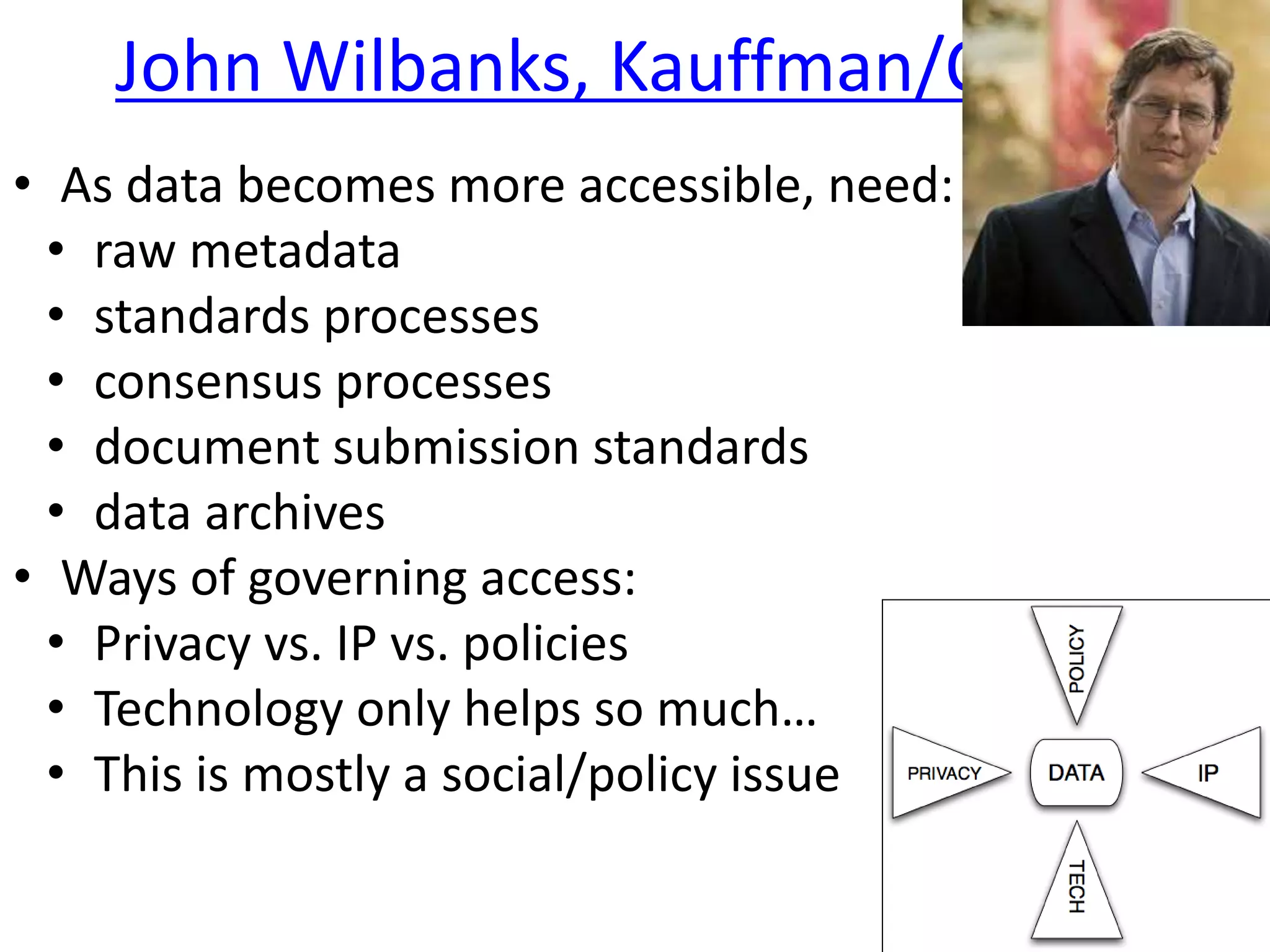
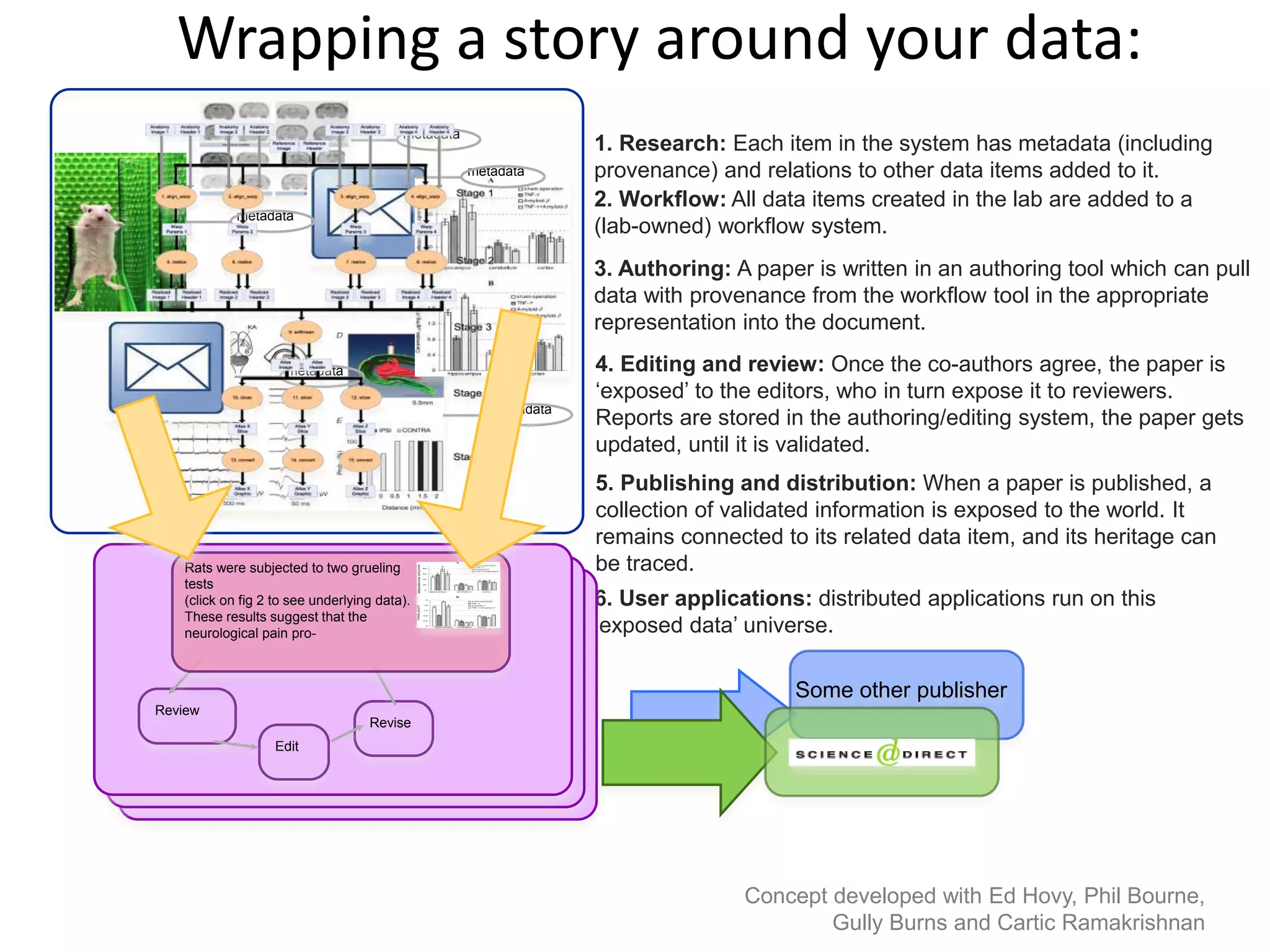
The document outlines key innovations and challenges in scholarly publishing, focusing on new publication formats, workflow integration, and alternative attribution models. It discusses various projects aimed at enhancing research accessibility and collaboration, including claim-evidence networks and executable papers. The future of scholarly communication emphasizes the importance of data sharing, citizen science, and the evolving role of publishers and libraries in this new landscape.











![“Publishers have been thinking we’re going out of
business for 20 years, what has suddenly changed?”
The internet! Not the technical web, but the social web….
‘The value of a […] network is proportional to the square of
the number of users of the system (n²)’ Metcalfe’s Law
1990’s:
Big Player
2000’s:
Medium Participant
2015:
Irrelevant!](https://image.slidesharecdn.com/force12-120406194021-phpapp02/75/How-to-Execute-A-Research-Paper-18-2048.jpg)
![19
What do we need?
[[1] Bleecker, J. ‘A Manifesto for Networked Objects — Cohabiting with Pigeons, Arphids and Aibos in the Internet of Things
http://nearfuturelaboratory.com/2006/02/26/a-manifesto-for-networked-objects/
2] Bechhofer, S., De Roure, D., Gamble, M., Goble, C. and Buchan, I. (2010) Research Objects: Towards Exchange and
Reuse of Digital Knowledge. In: The Future of the Web for Collaborative Science (FWCS 2010), April 2010, Raleigh, NC, USA.
http://precedings.nature.com/documents/4626/version/1
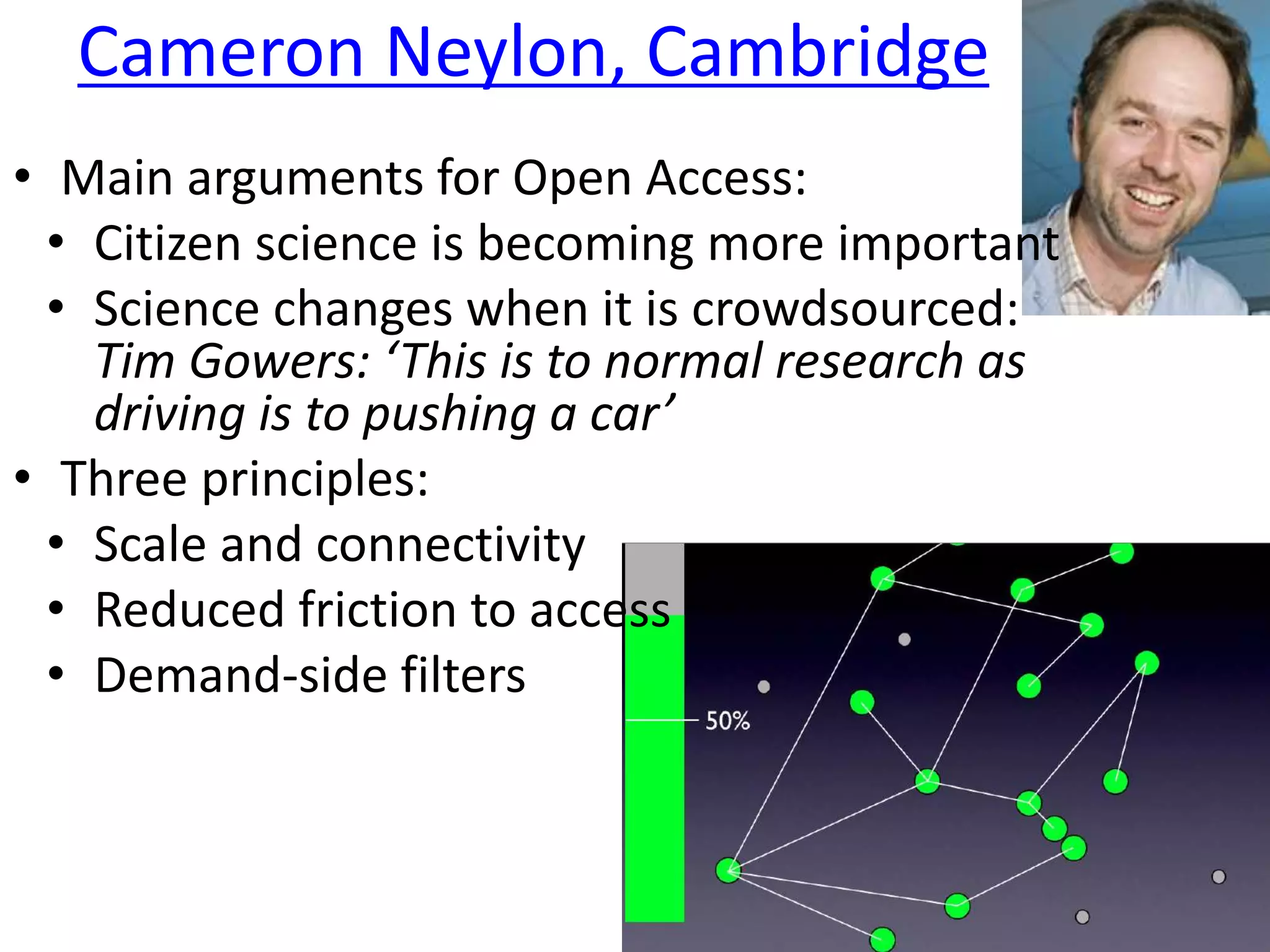
[3] Neylon, C. ‘Network Enabled Research: Maximise scale and connectivity, minimise friction’,
http://cameronneylon.net/blog/network-enabled-research/ ‘
Internet of things: (Bleecker, [1])
Interact with ‘objects that blog’ or ‘Blogjects’, that:
track where they are and where they’ve been;have
histories of their encounters and experienceshave
agency - an assertive voice on the social web [2]
Research Objects: (Bechofer et al, [2])
Create semantically rich aggregations of resources,
that can possess some scientific intent or support
some research objective
Networked Knowledge: (Neylon, [3])
If we care about taking advantage of the web and
internet for research then we must tackle the building
of scholarly communication networks.
These networks will have two critical characteristics:
scale and a lack of friction. [3]](https://image.slidesharecdn.com/force12-120406194021-phpapp02/75/How-to-Execute-A-Research-Paper-19-2048.jpg)



![How a claim becomes a fact:
• Voorhoeve, 2006: “These miRNAs neutralize p53- mediated CDK
inhibition, possibly through direct inhibition of the expression of the
tumorsuppressor LATS2.”
• Kloosterman and Plasterk, 2006: “In a genetic screen, miR-372 and miR-
373 were found to allow proliferation of primary human cells that
express oncogenic RAS and active p53, possibly by inhibiting the tumor
suppressor LATS2 (Voorhoeve et al., 2006).”
• Yabuta et al., 2007: “[On the other hand,] two miRNAs, miRNA-372 and-
373, function as potential novel oncogenes in testicular germ cell
tumors by inhibition of LATS2 expression, which suggests that Lats2 is an
important tumor suppressor (Voorhoeve et al., 2006).”
• Okada et al., 2011: “Two oncogenic miRNAs, miR-372 and miR-373,
directly inhibit the expression of Lats2, thereby allowing tumorigenic
growth in the presence of p53 (Voorhoeve et al., 2006).”](https://image.slidesharecdn.com/force12-120406194021-phpapp02/75/How-to-Execute-A-Research-Paper-24-2048.jpg)
![Working on ontology:
1. Add to formal knowledge representations, e.g. Biological
Expression Language add {V = 3, S = N, B = 0}:
• SET Evidence = "Arterial cells are highly susceptible to oxidative stress, which can induce
both necrosis and apoptosis (programmed cell death) [1,2]"
• biologicalProcess(GO:"response to oxidative stress") increases
biologicalProcess(GO:"apoptotic process")
• biologicalProcess(GO:"response to oxidative stress") increases
biologicalProcess(GO:necrosis)
2. Improve triple search engines, e.g. compare in iHop:
• The Lats2 tumor suppressor protein has been implicated earlier in promoting p53
activation in response to mitotic apparatus stress {V = 2, S = NN, B = 0}
• Our findings reveal that miR-373 would be a potential oncogene and it participates in
the carcinogenesis of human esophageal cancer {V = 1/2?, S = A, B = D}](https://image.slidesharecdn.com/force12-120406194021-phpapp02/75/How-to-Execute-A-Research-Paper-25-2048.jpg)