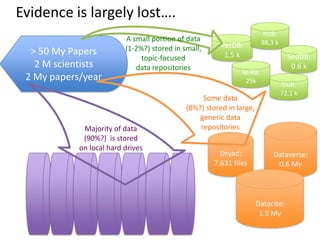
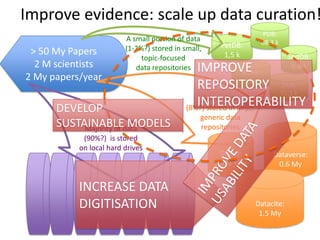
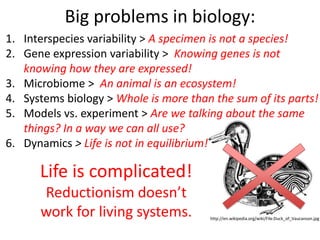
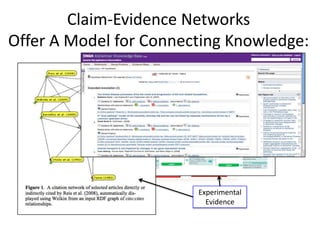
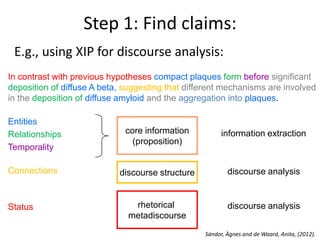
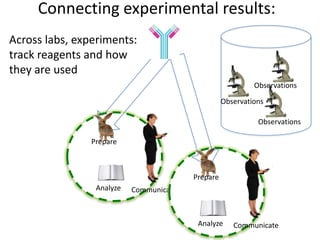
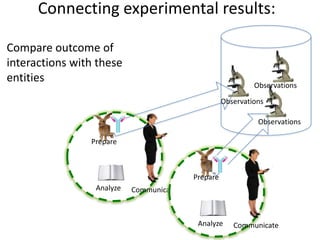
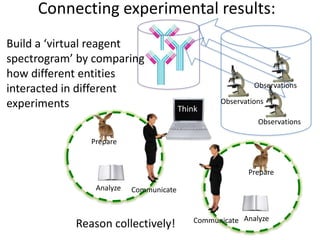
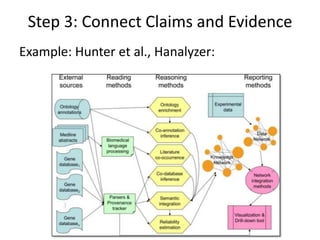
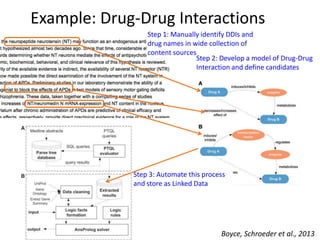
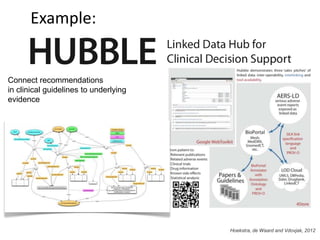
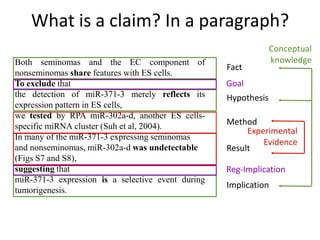
This document discusses connecting biological knowledge through claim-evidence networks. It outlines some of the challenges in biology like variability between specimens and gene expression changes. It then proposes that claim-evidence networks can be used to connect biological knowledge by linking experimental evidence to claims. Steps to build these networks include identifying claims in documents, structuring the evidence in databases, and automatically connecting the claims and evidence. Examples of efforts that link drug interactions to evidence and predict protein interactions across species are provided. However, it notes that more still needs to be done to fully realize this approach.












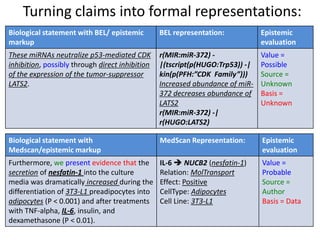






![• Voorhoeve et al., 2006: “These miRNAs neutralize p53- mediated CDK
inhibition, possibly through direct inhibition of the expression of the tumor
suppressor LATS2.”
• Kloosterman and Plasterk, 2006: “In a genetic screen, miR-372 and miR-373
were found to allow proliferation of primary human cells that express
oncogenic RAS and active p53, possibly by inhibiting the tumor suppressor
LATS2 (Voorhoeve et al., 2006).”
• Okada et al., 2011: “Two oncogenic miRNAs, miR-372 and miR-373, directly
inhibit the expression of Lats2, thereby allowing tumorigenic growth in the
presence of p53 (Voorhoeve et al., 2006).”
“[Y]ou can transform .. fiction into fact, just by adding
or subtracting references”, Latour, 1987
What is the claim? Who makes it?](https://image.slidesharecdn.com/ismbdewaard-130723121627-phpapp01/85/Why-Life-is-Difficult-and-What-We-MIght-Do-About-It-32-320.jpg)