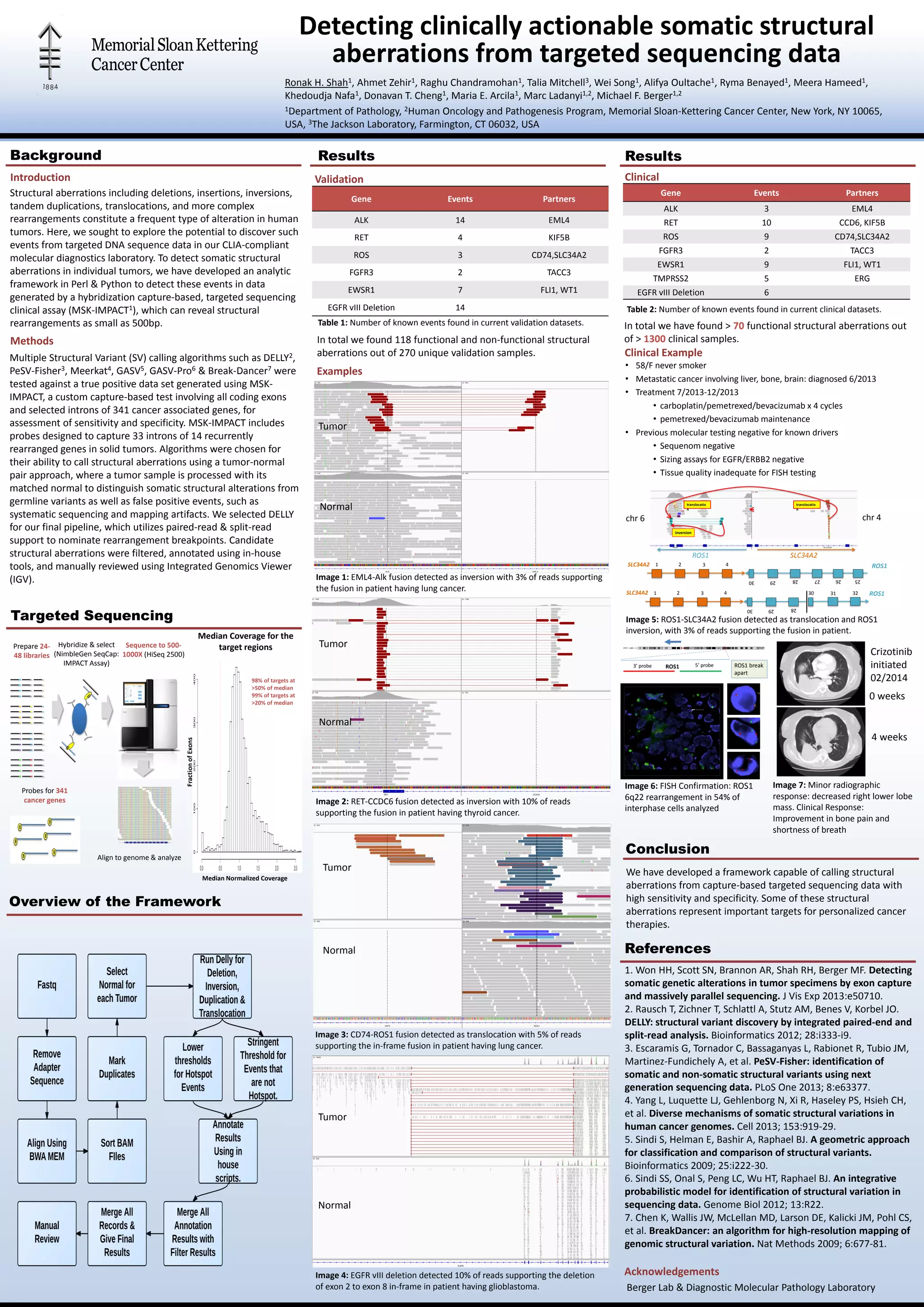
The document presents a framework developed to detect clinically actionable somatic structural aberrations from targeted sequencing data, utilizing algorithms like delly for high sensitivity and specificity. The study highlights the ability to identify various structural genetic alterations in tumors, which could serve as targets for personalized cancer therapies. A significant number of functional structural aberrations were found across numerous clinical samples, demonstrating the potential of the method in enhancing molecular diagnostics.