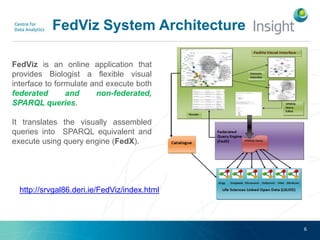
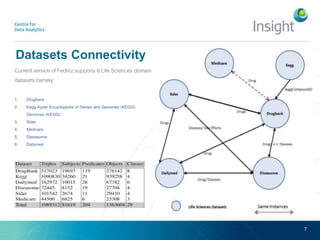
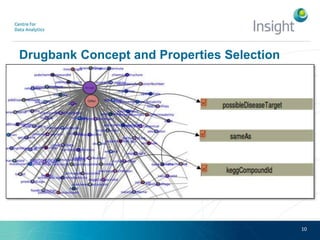
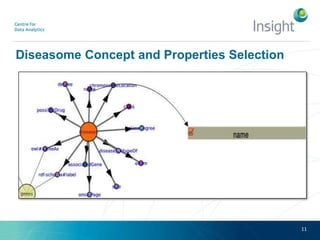
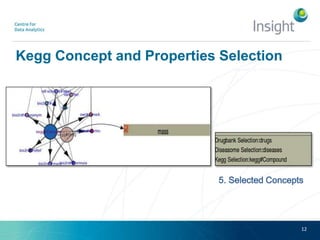
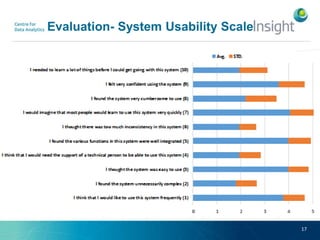
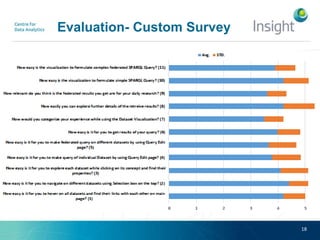
Fedviz is an online application designed to aid biologists in formulating and executing federated and non-federated SPARQL queries through a visual interface. It supports six life sciences datasets and aims to simplify the complex process of querying multiple data sources related to drug interactions and diseases. An evaluation indicated a high usability score of 74.16%, suggesting that users found it intuitive and effective for complex query formulation.