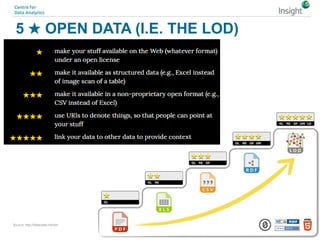
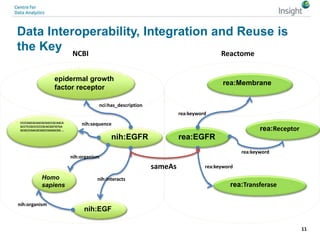
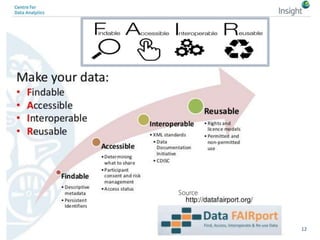
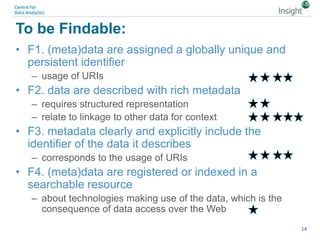
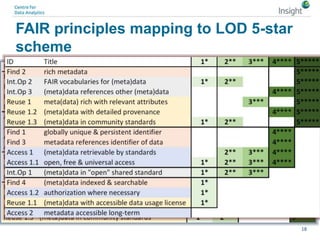
The document discusses the comparison between FAIR data principles and the 5-star open data principles, focusing on aspects like interoperability and findability. It highlights the distinctions and overlaps between the two frameworks, emphasizing that FAIR principles prioritize user needs and data reusability, while 5-star principles offer a more idealistic approach to open data. The document concludes by questioning the future effectiveness of both principles regarding data reusability and accessibility.