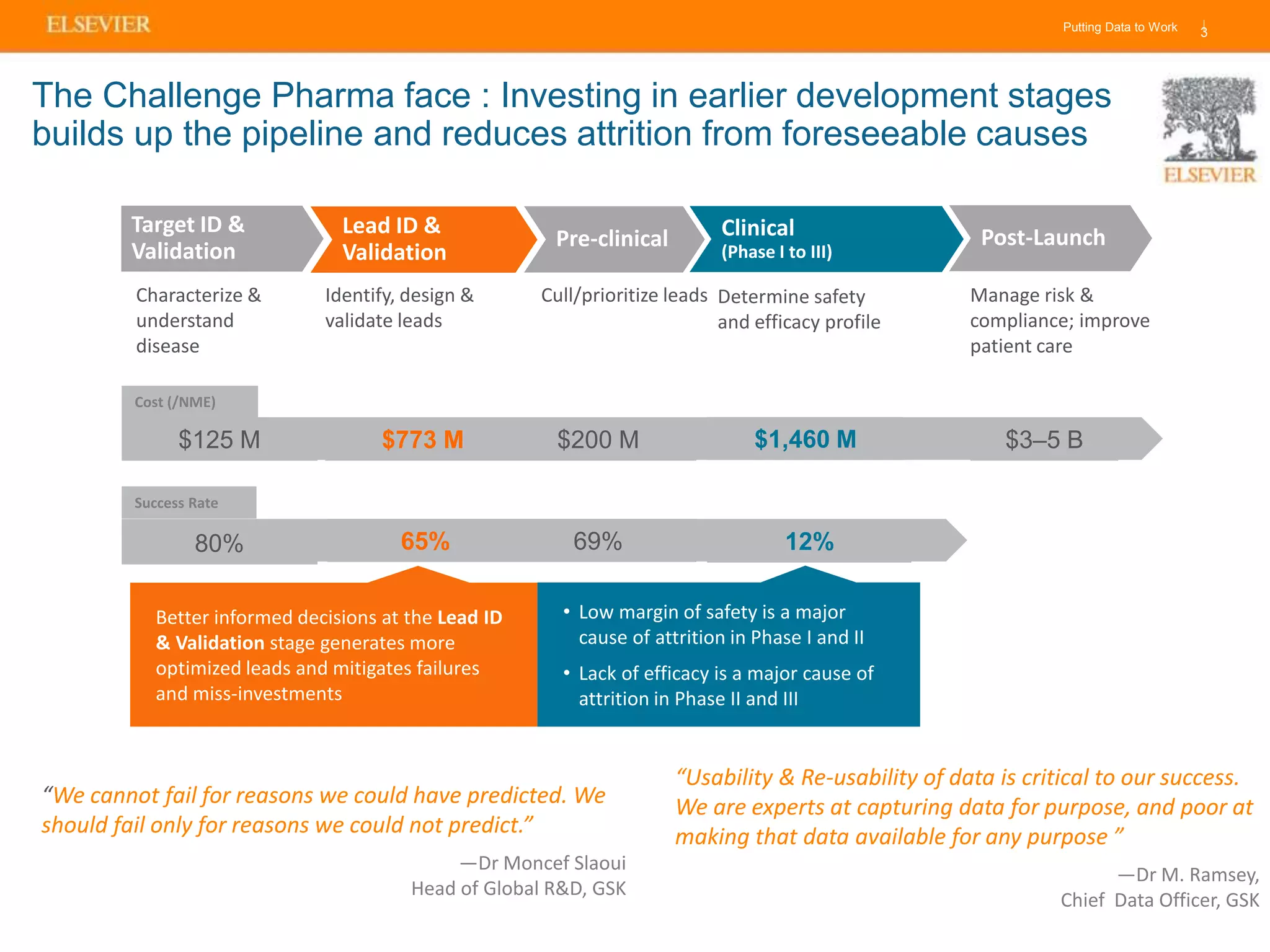
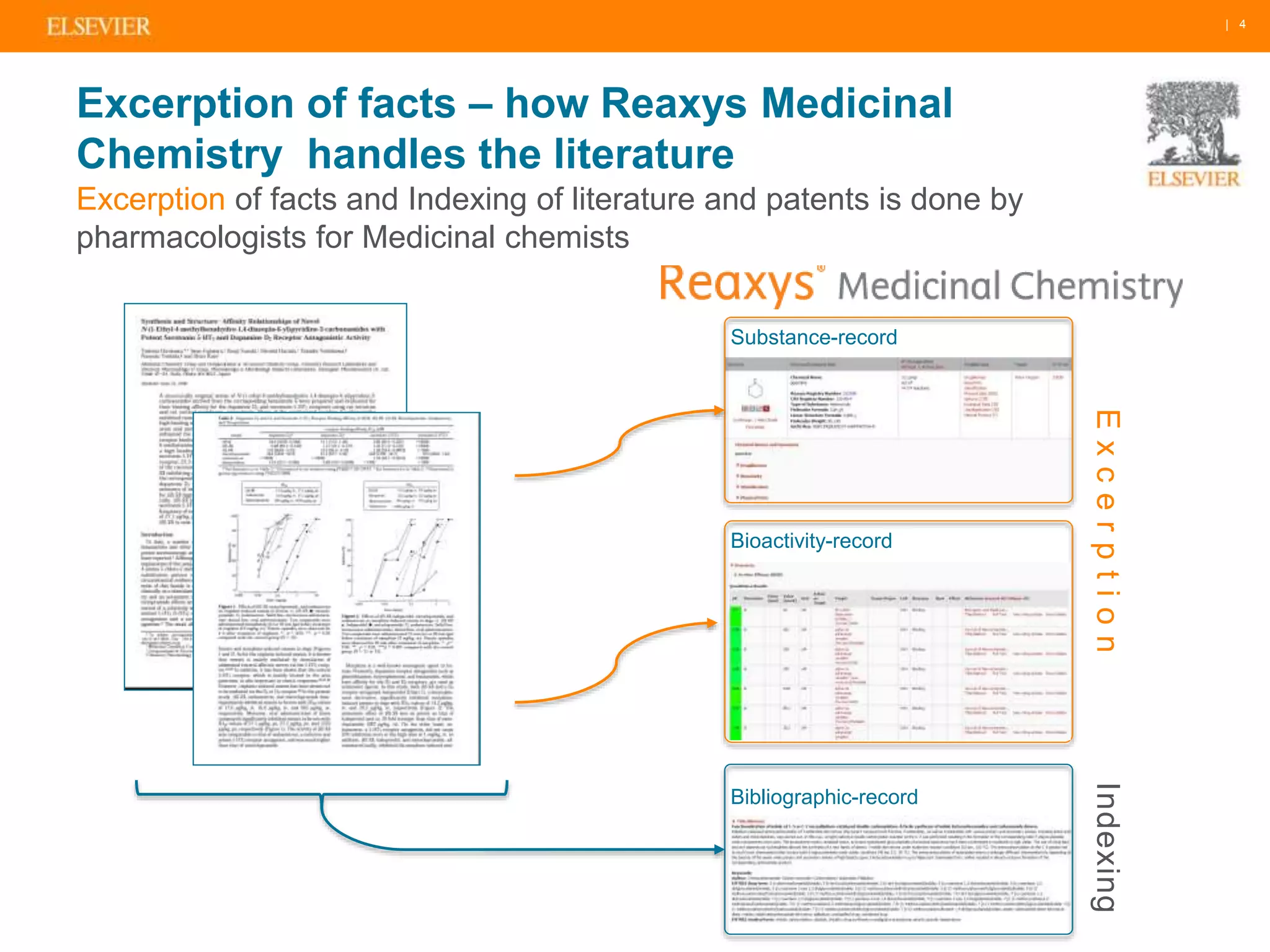
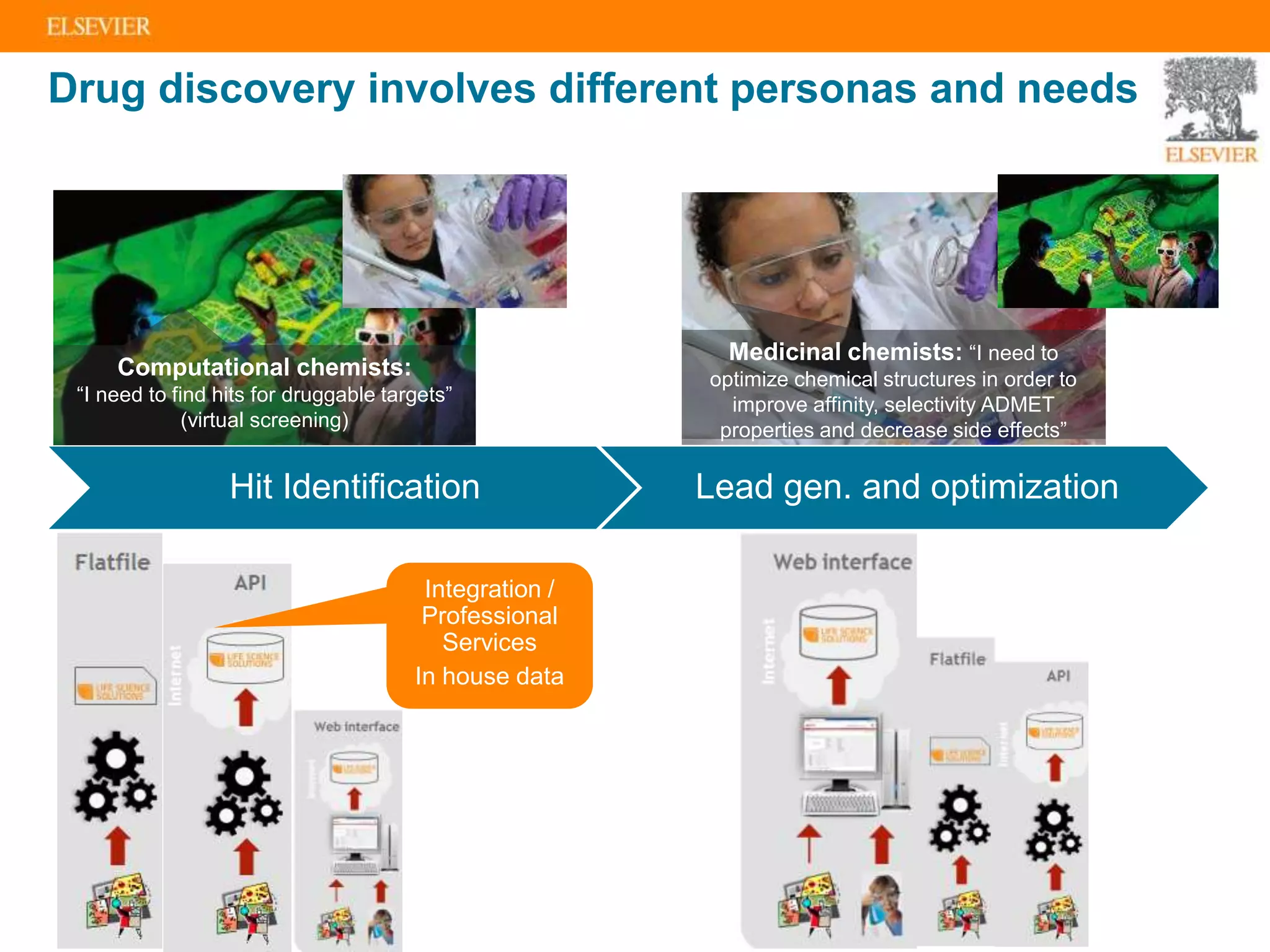
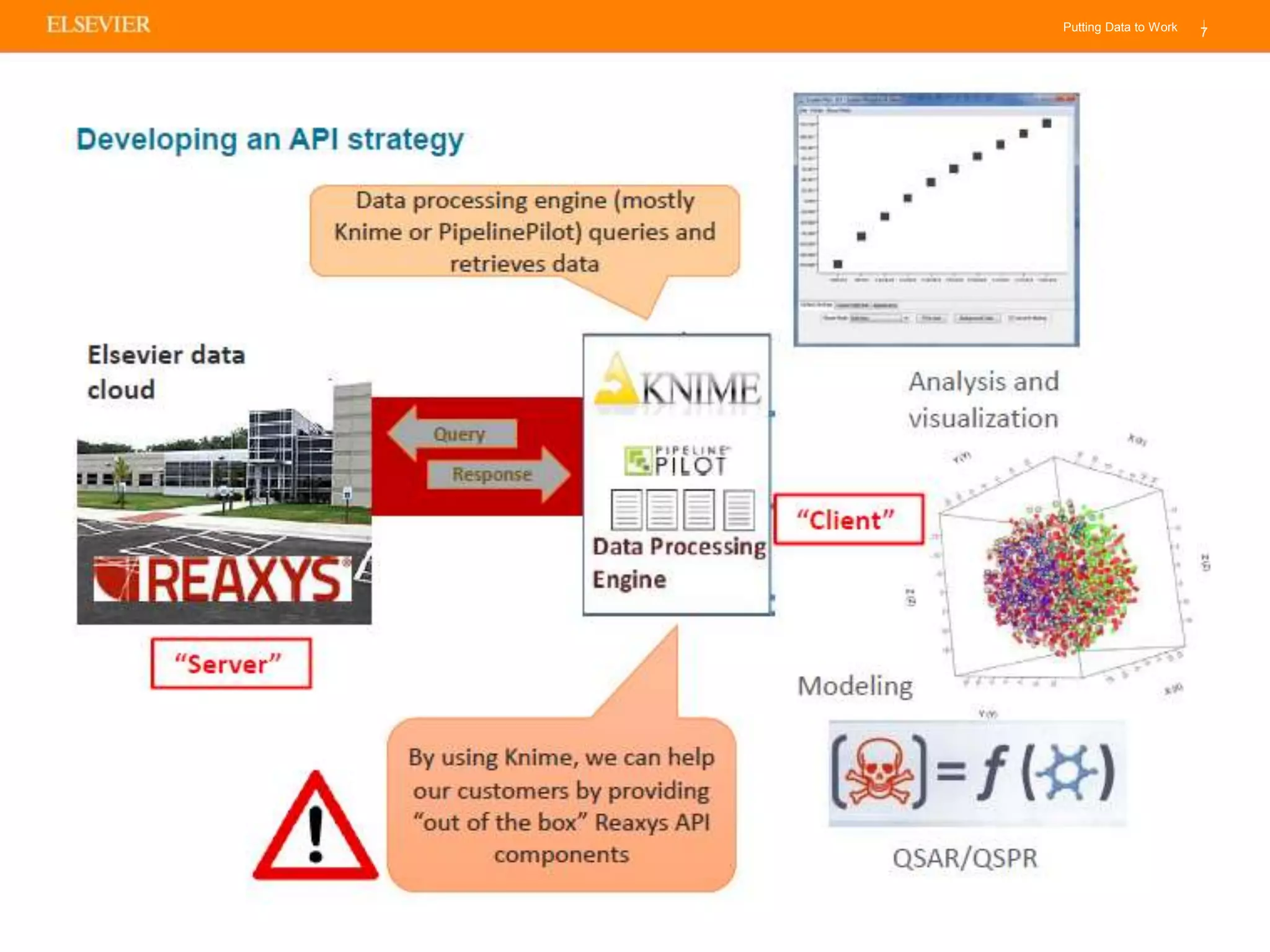
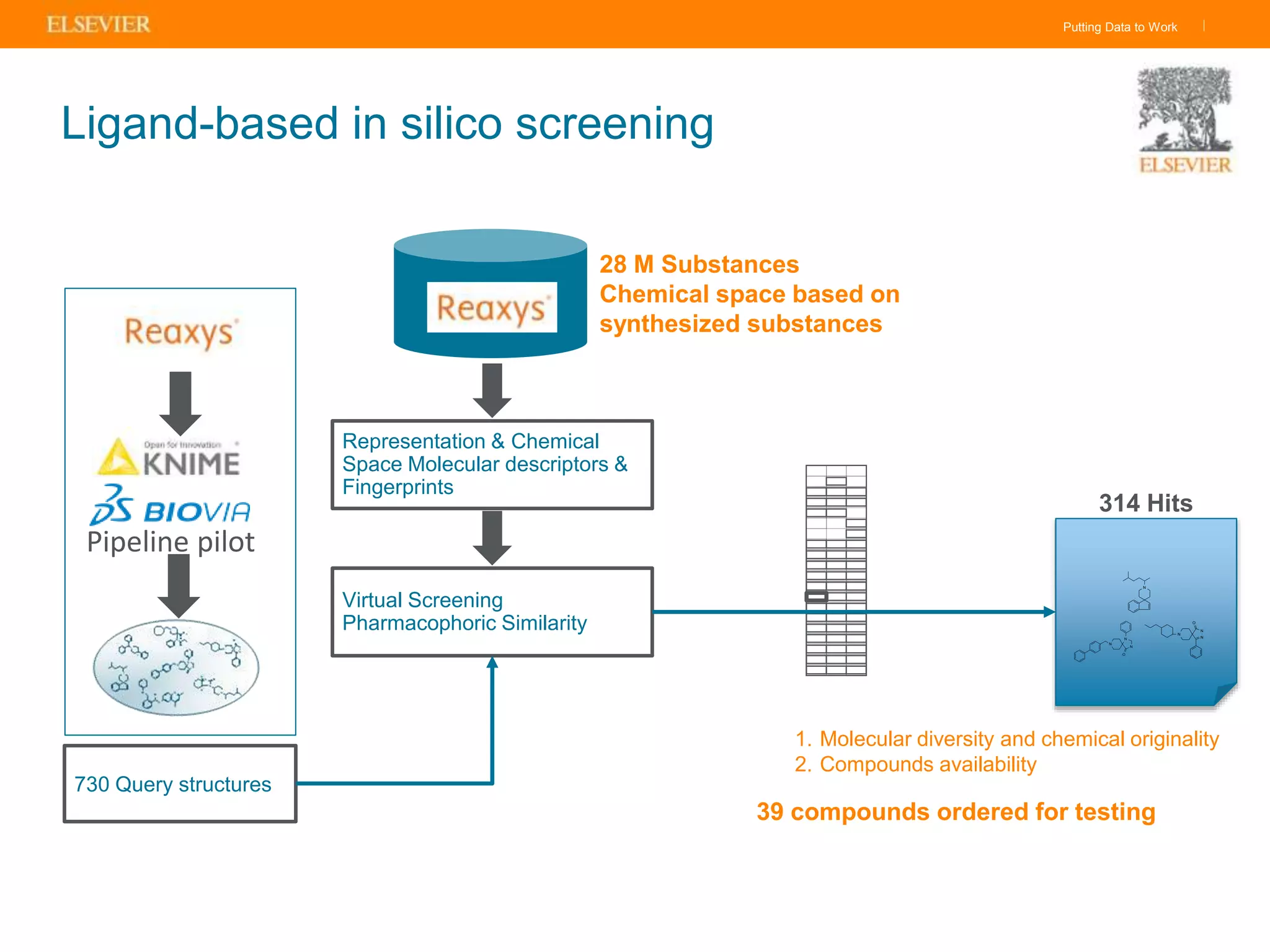
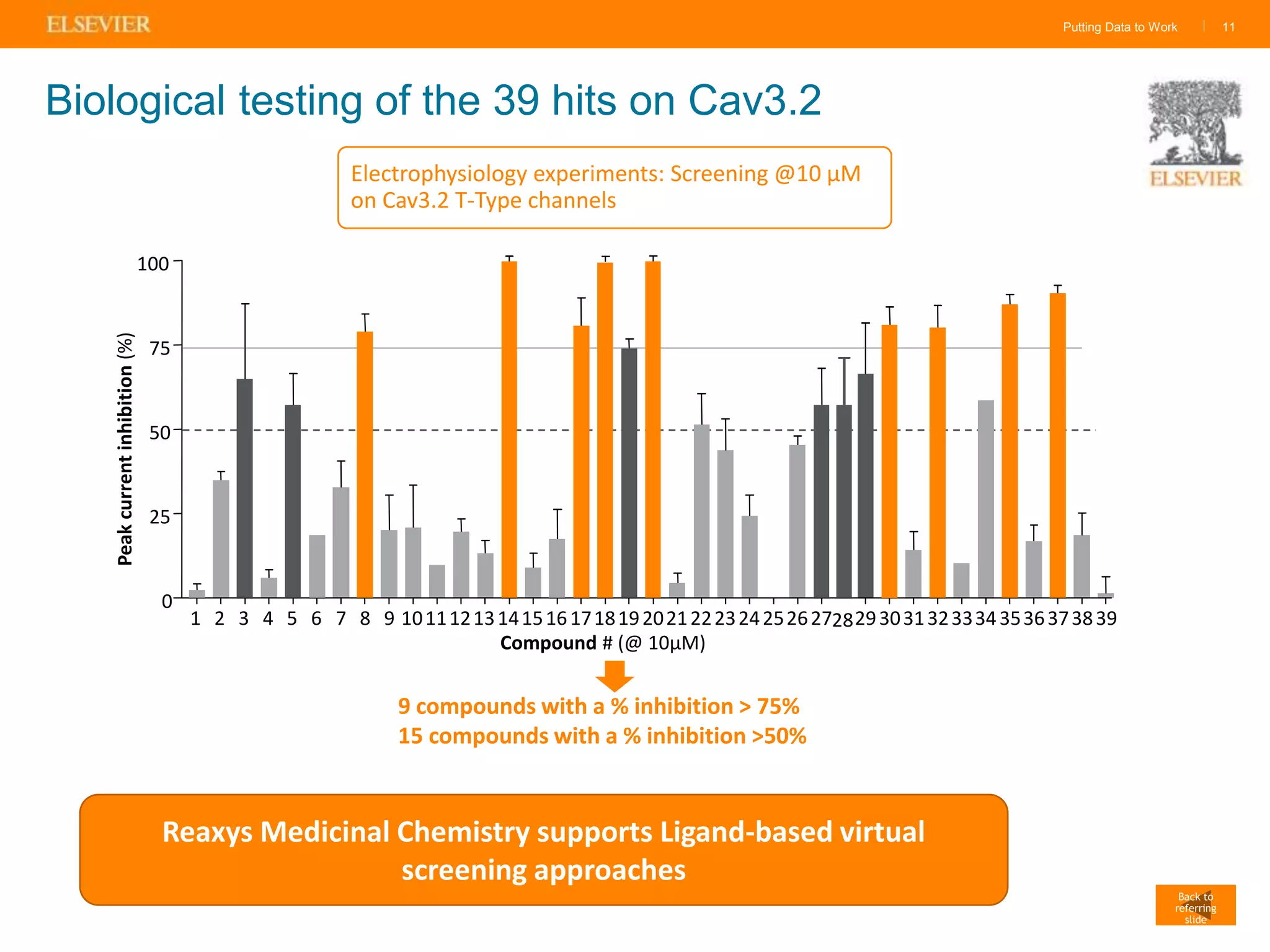
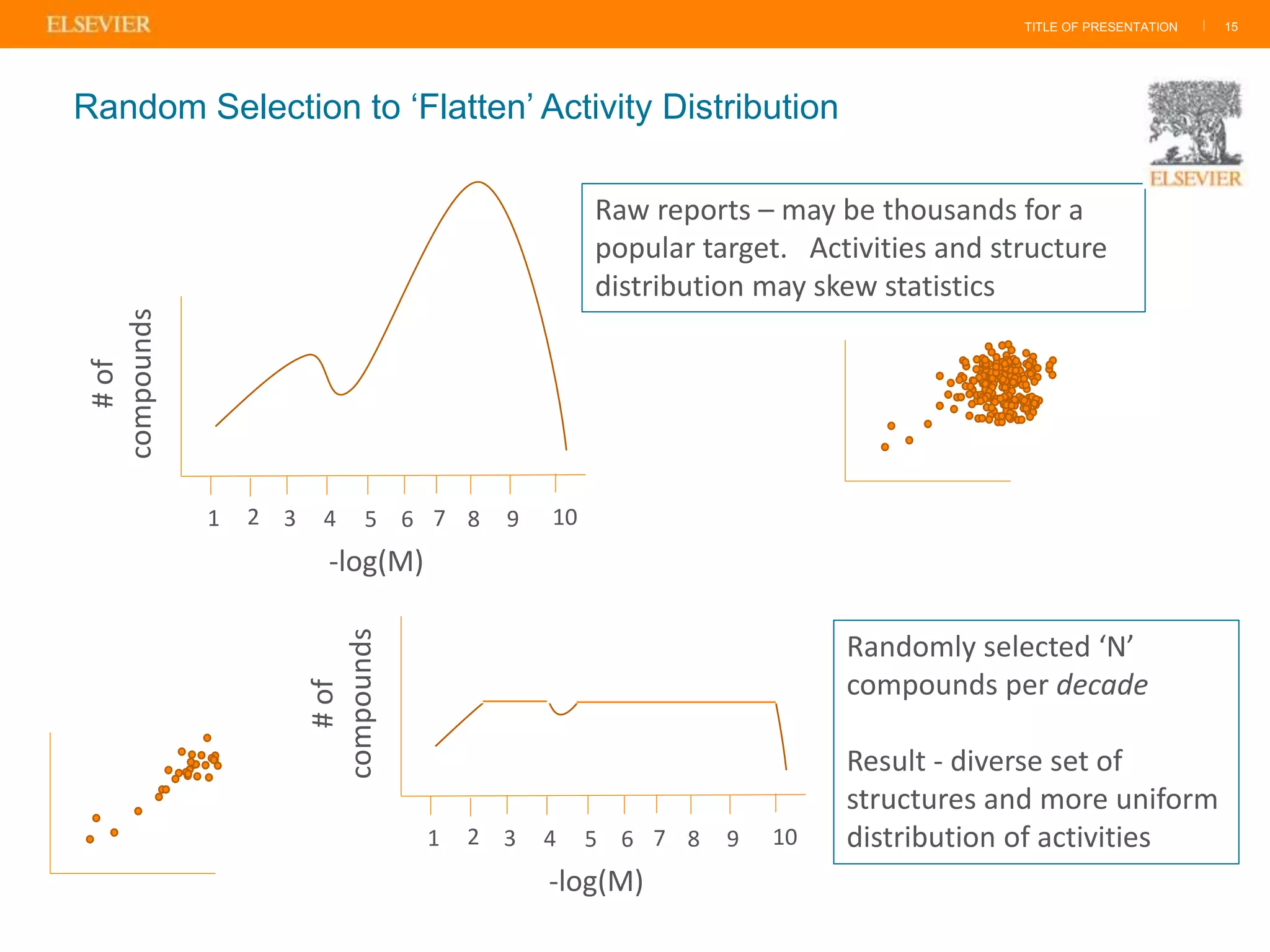
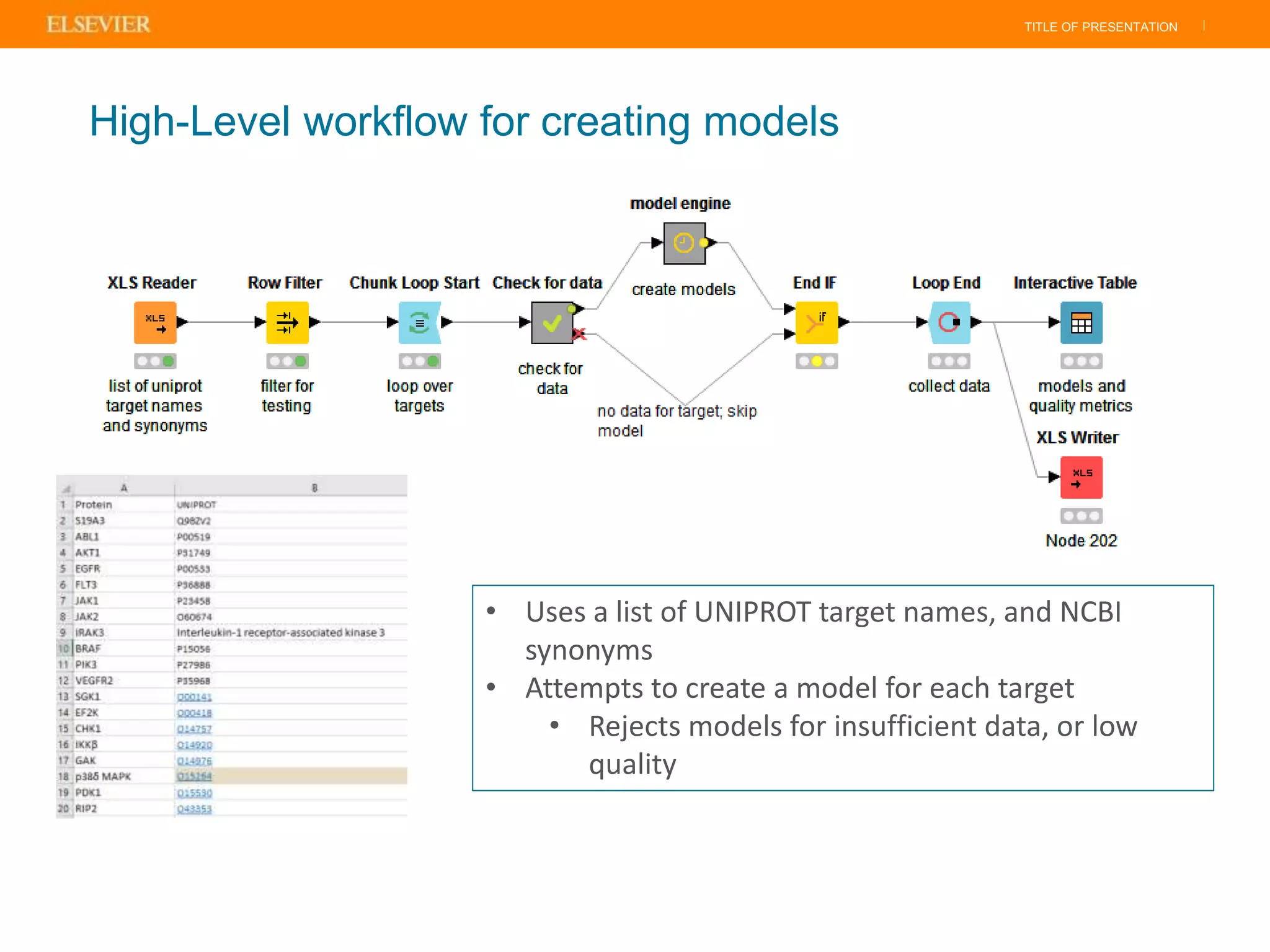
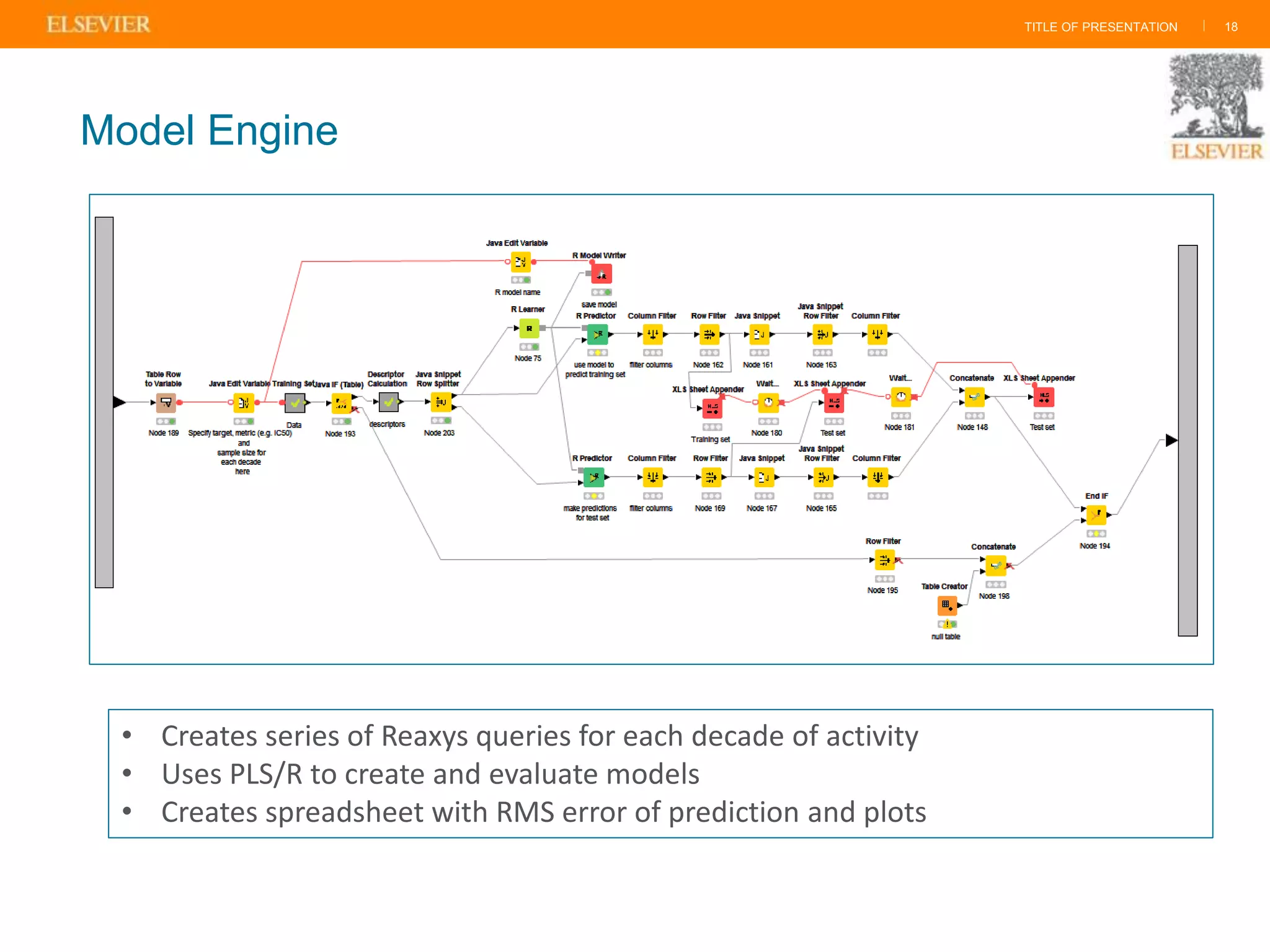
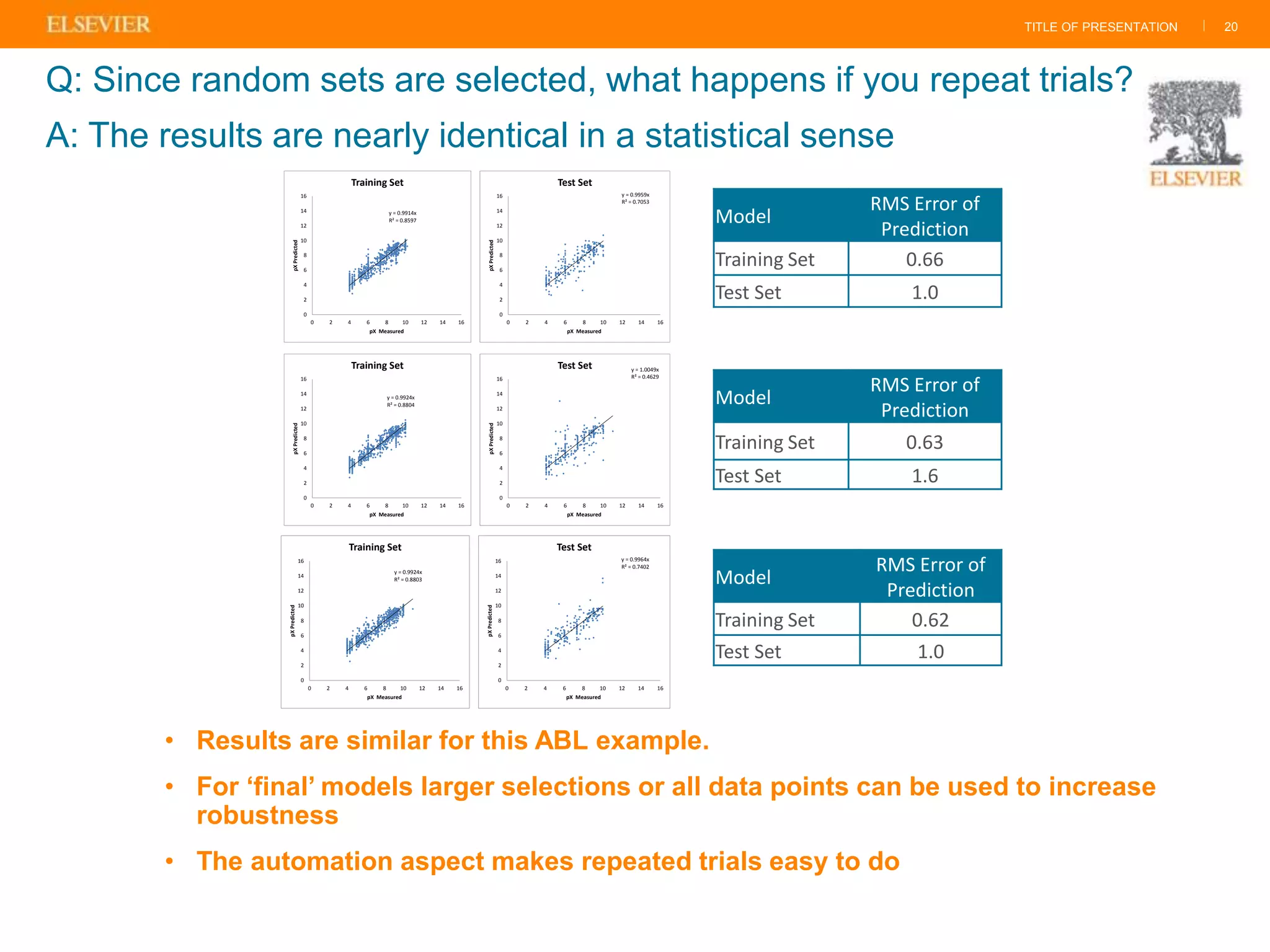
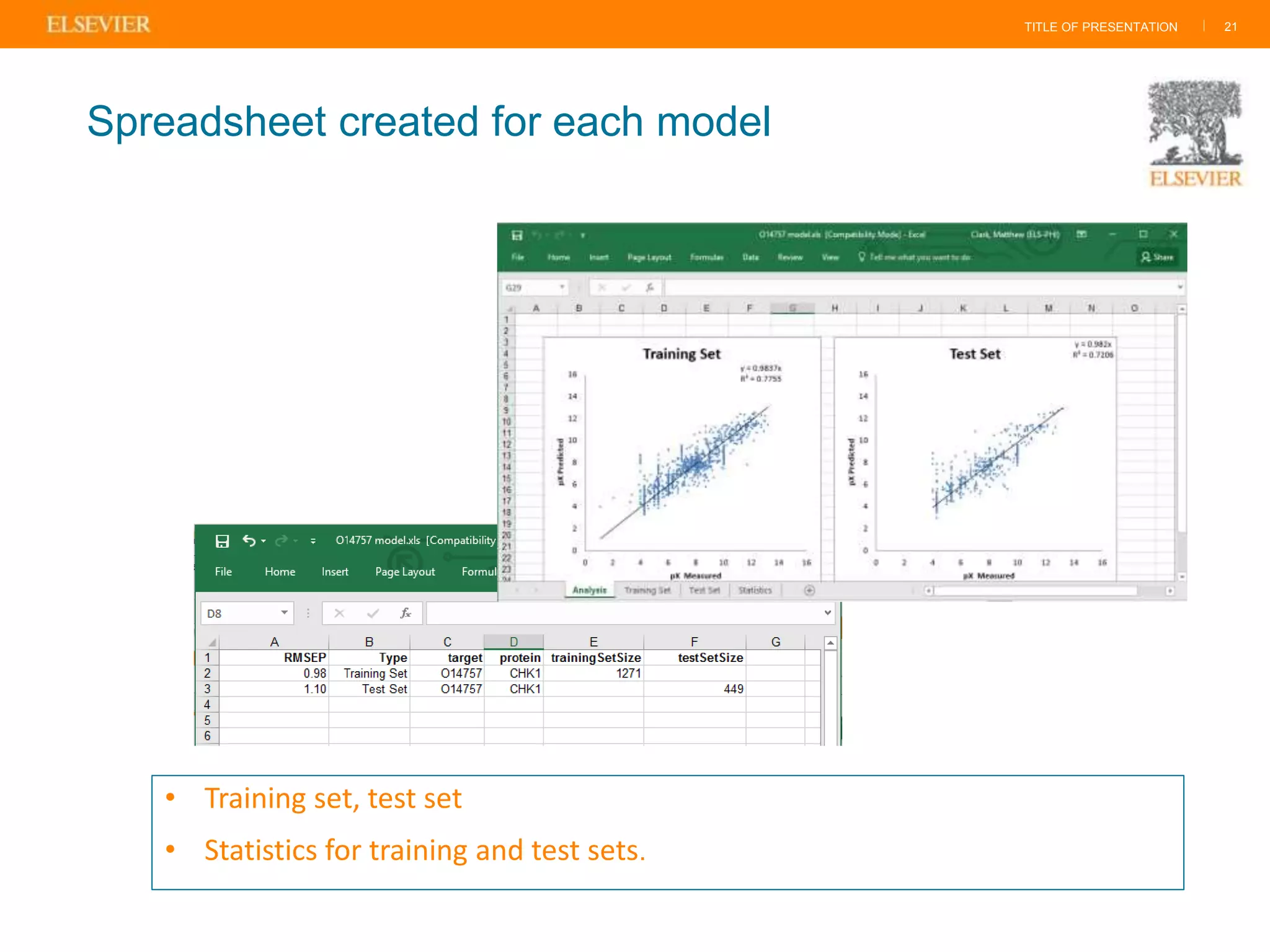
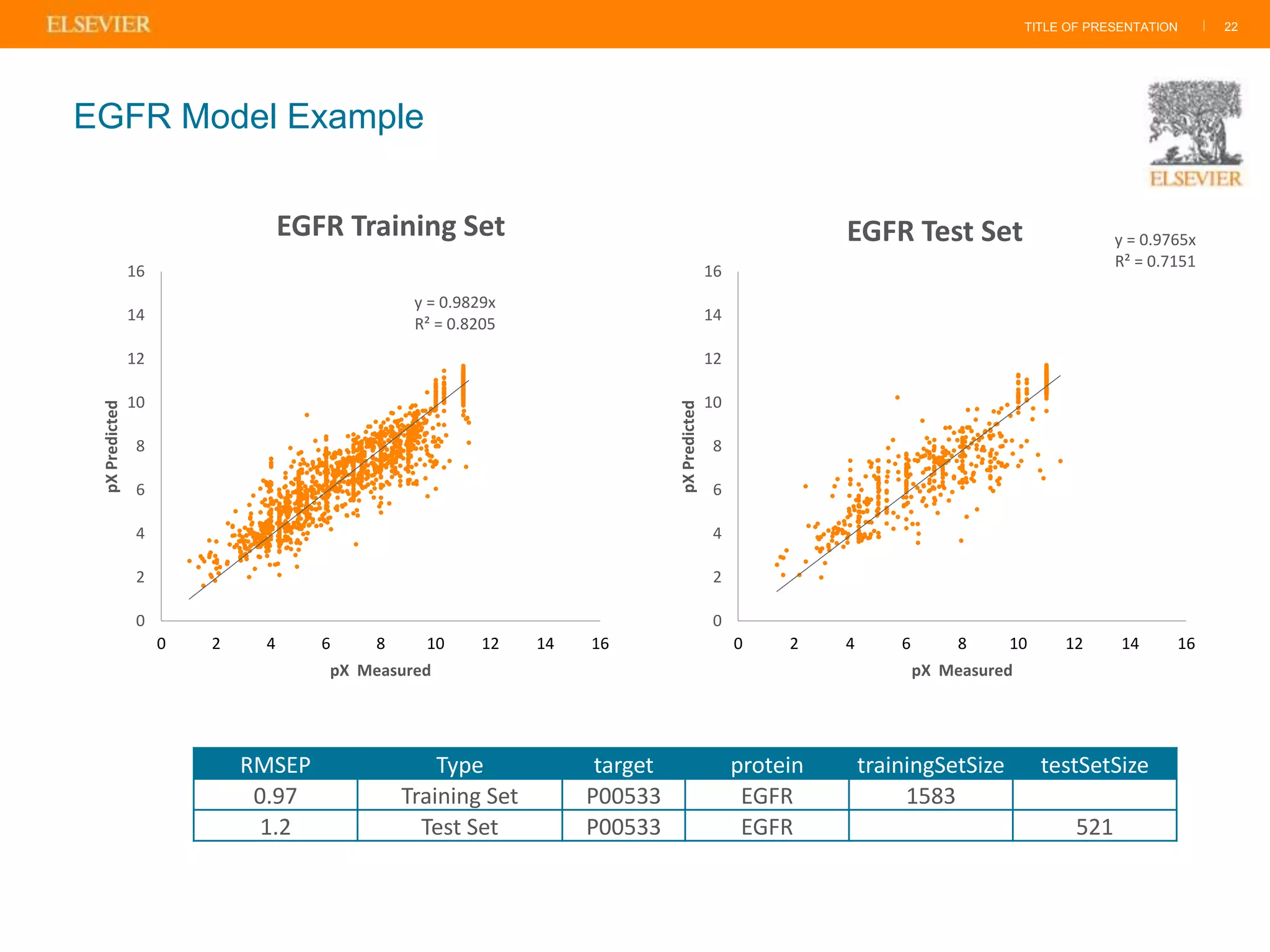
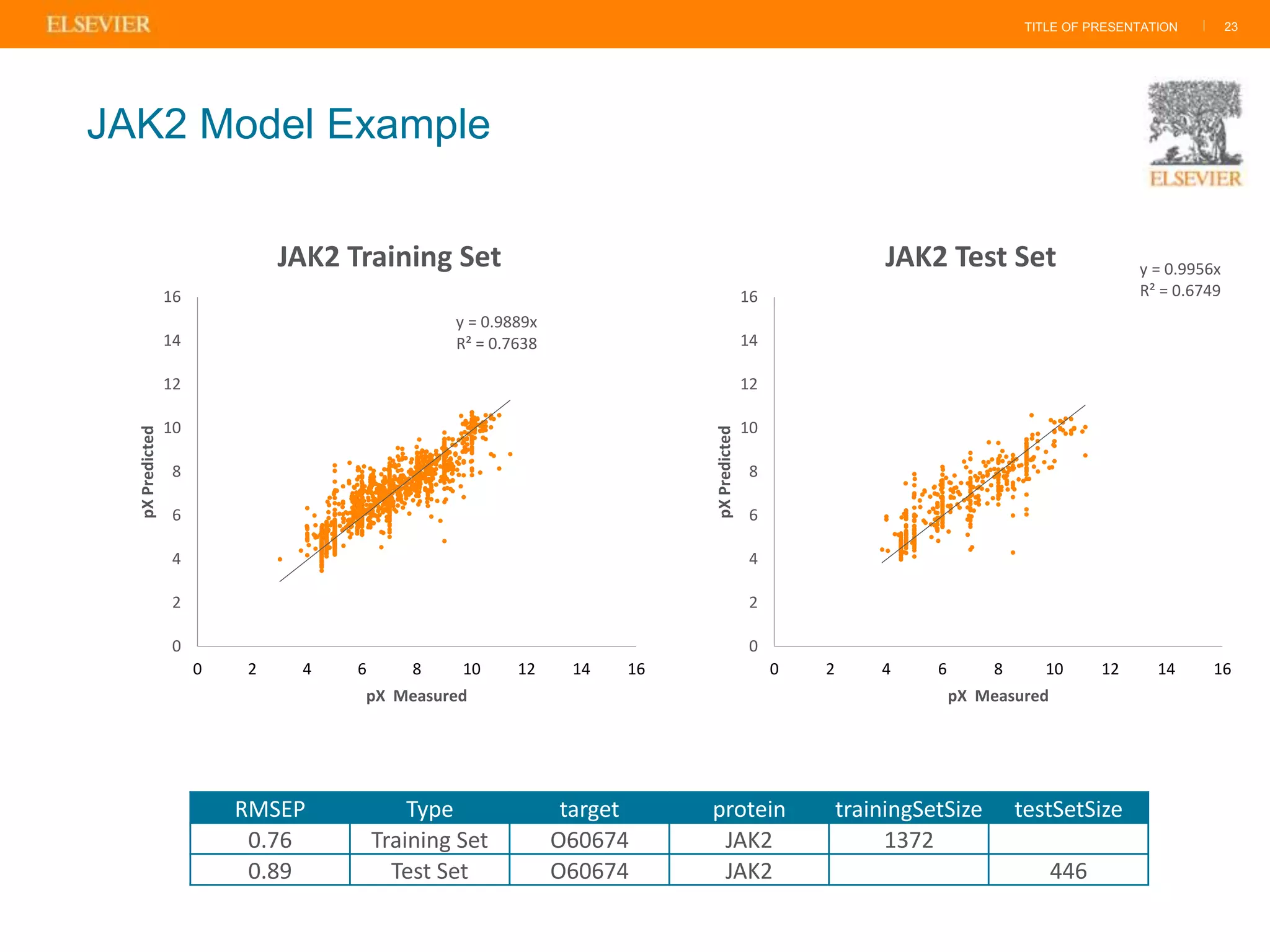
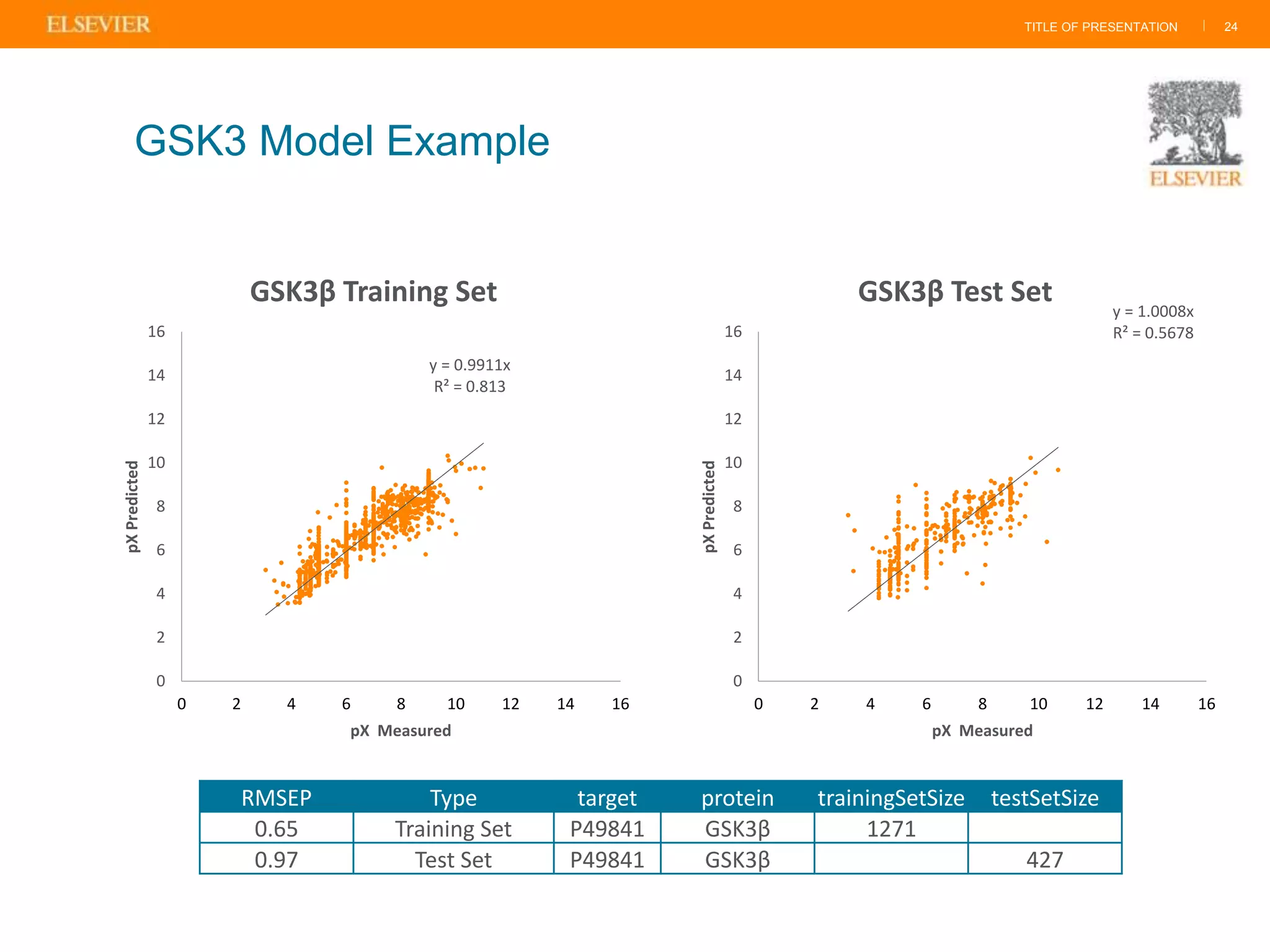
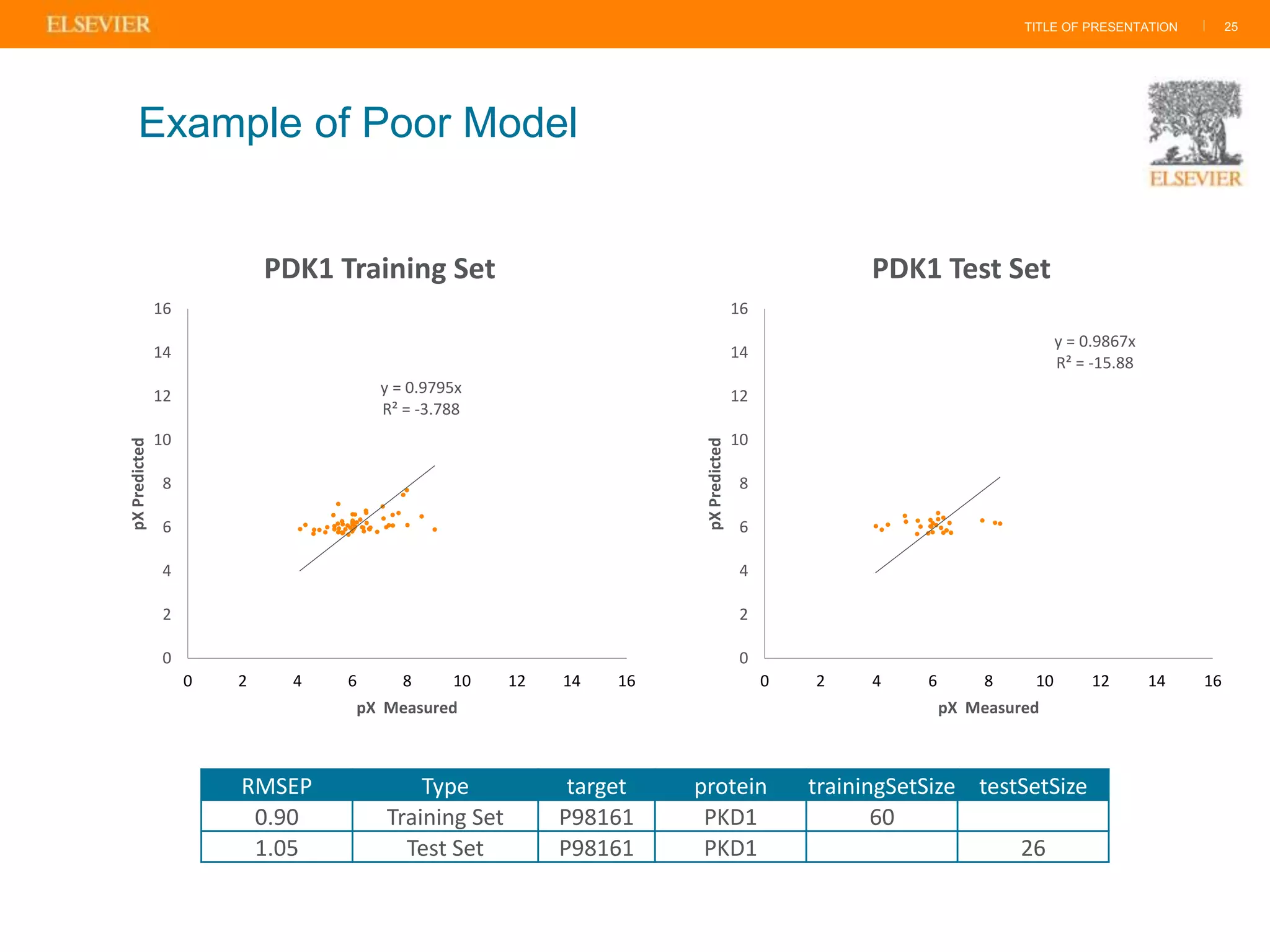
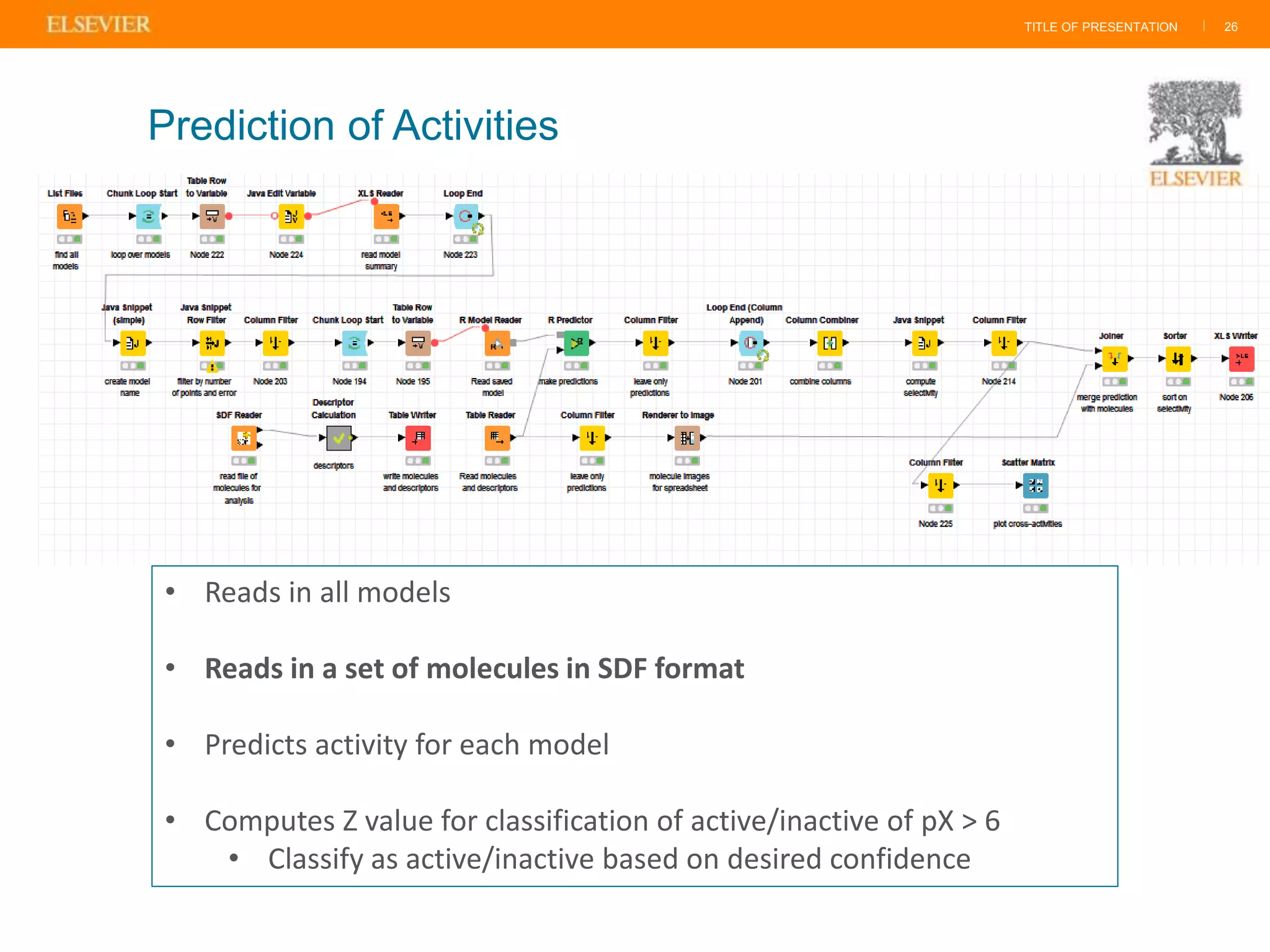
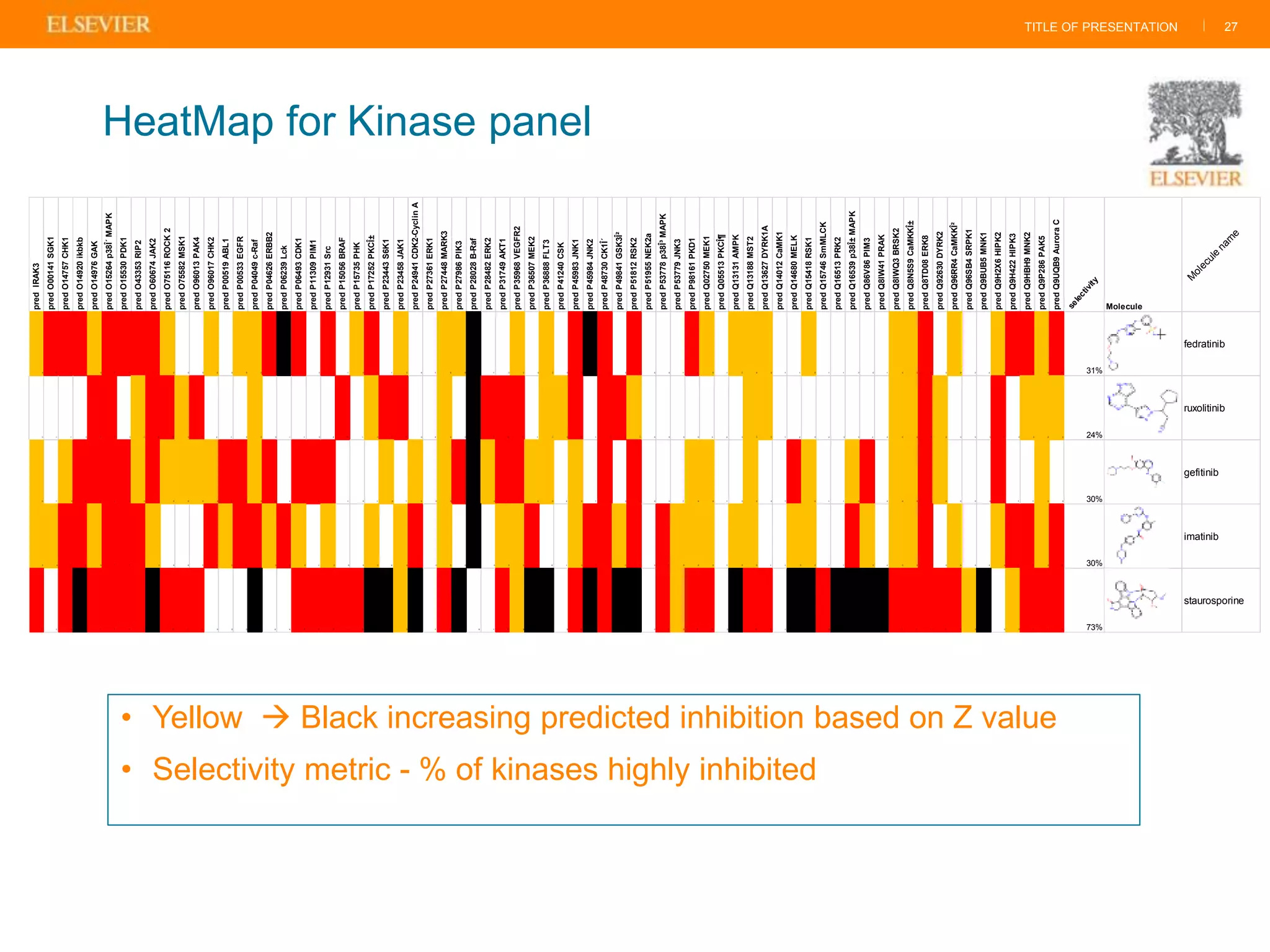
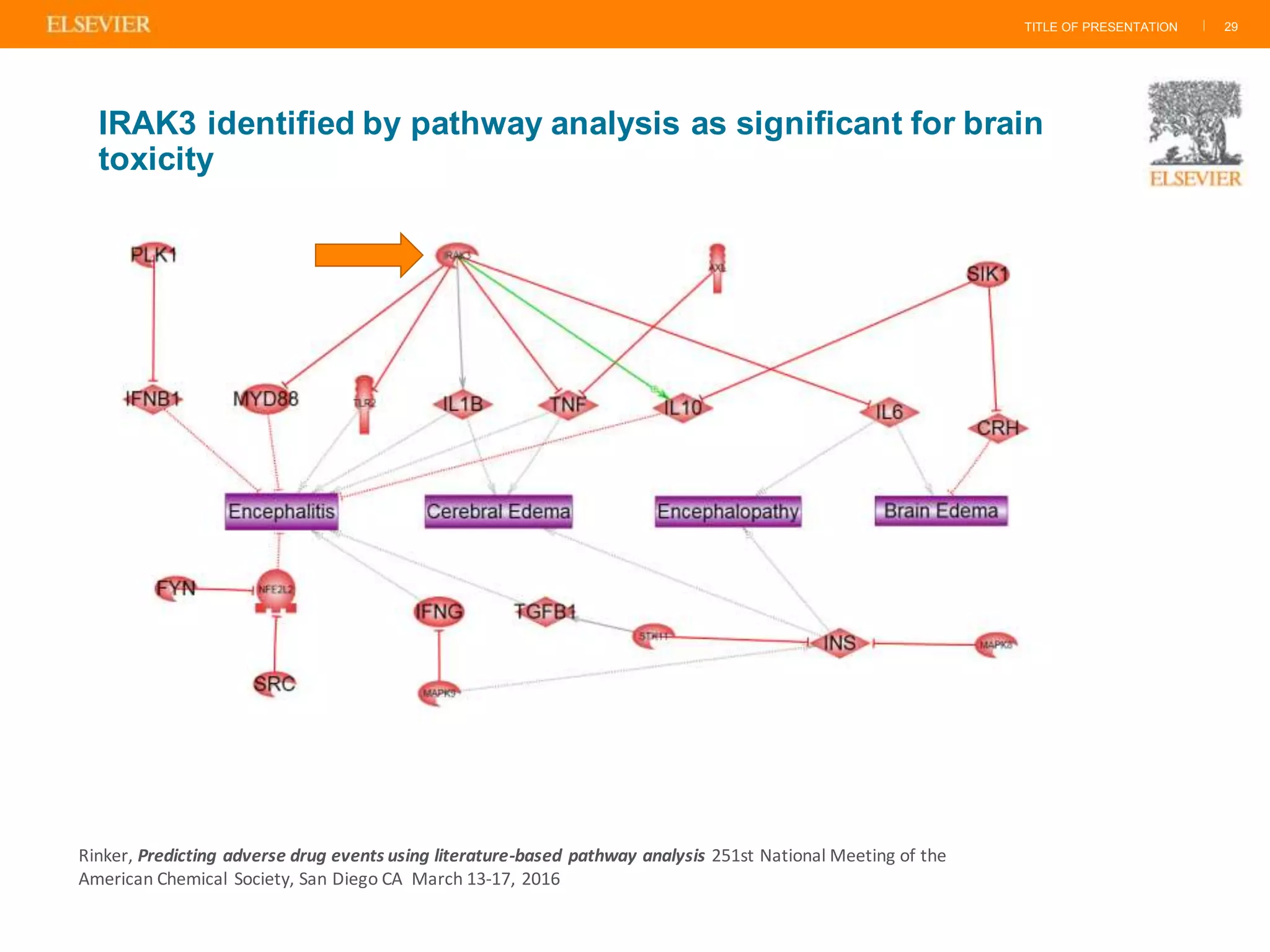
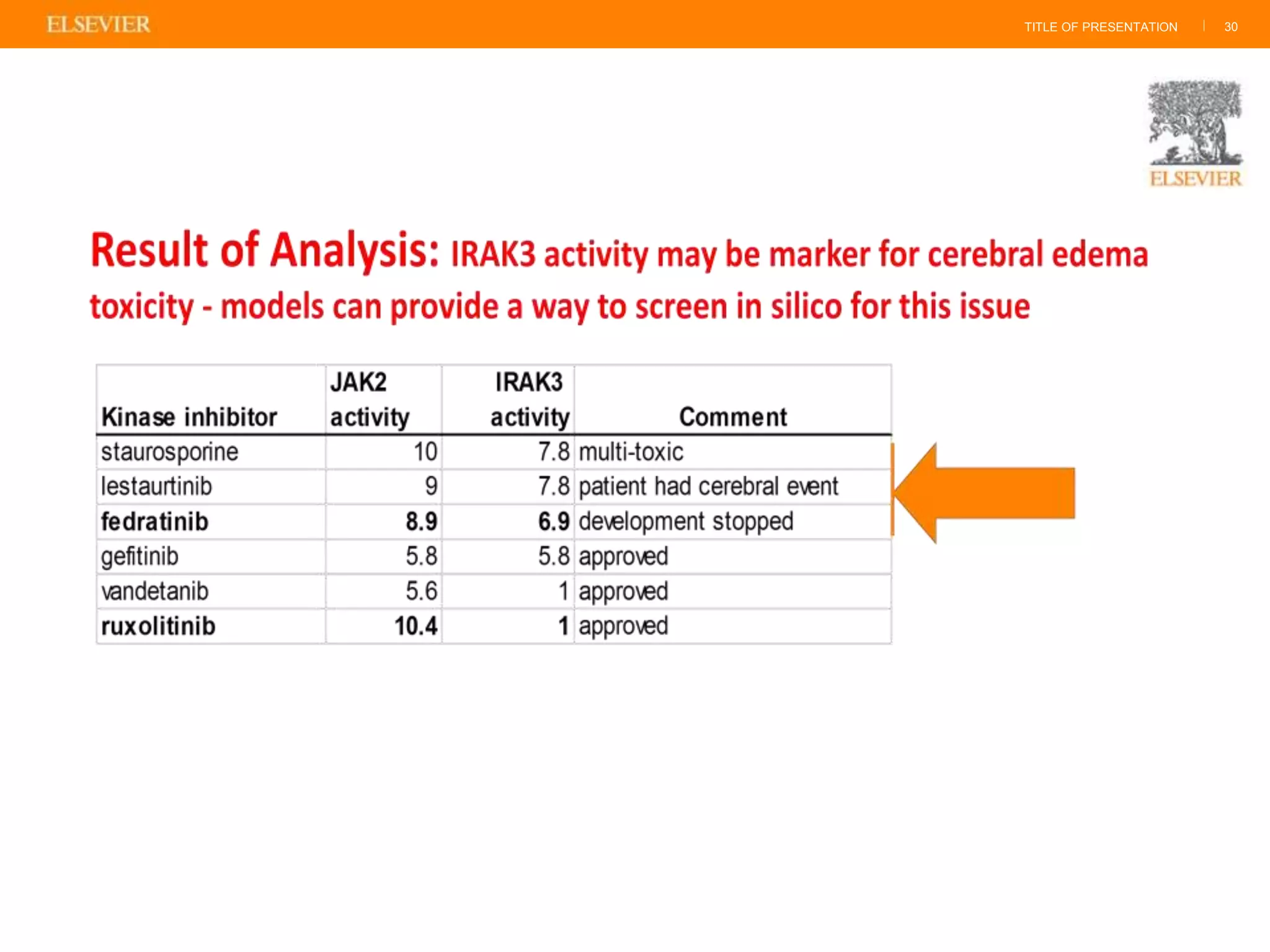
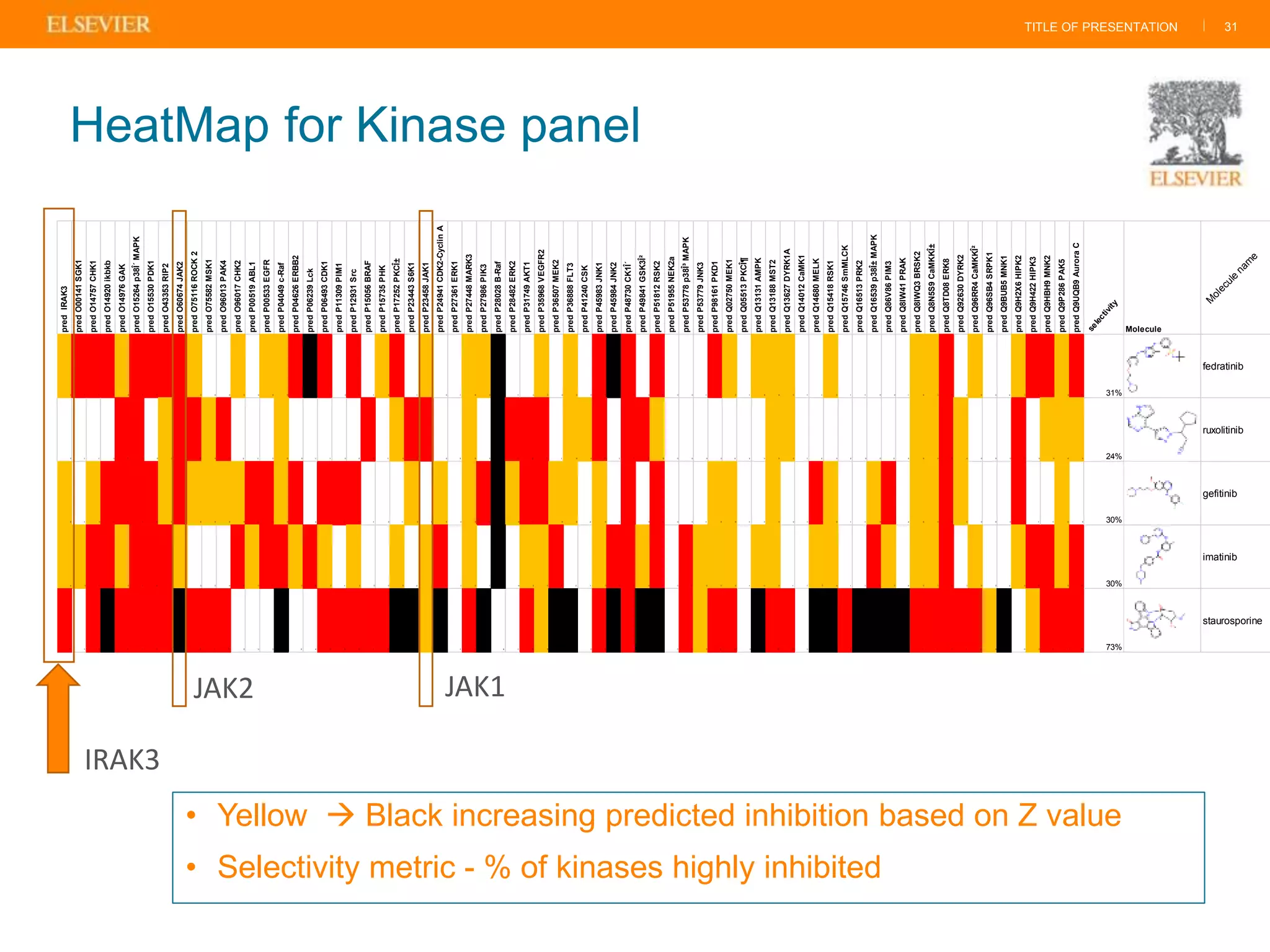
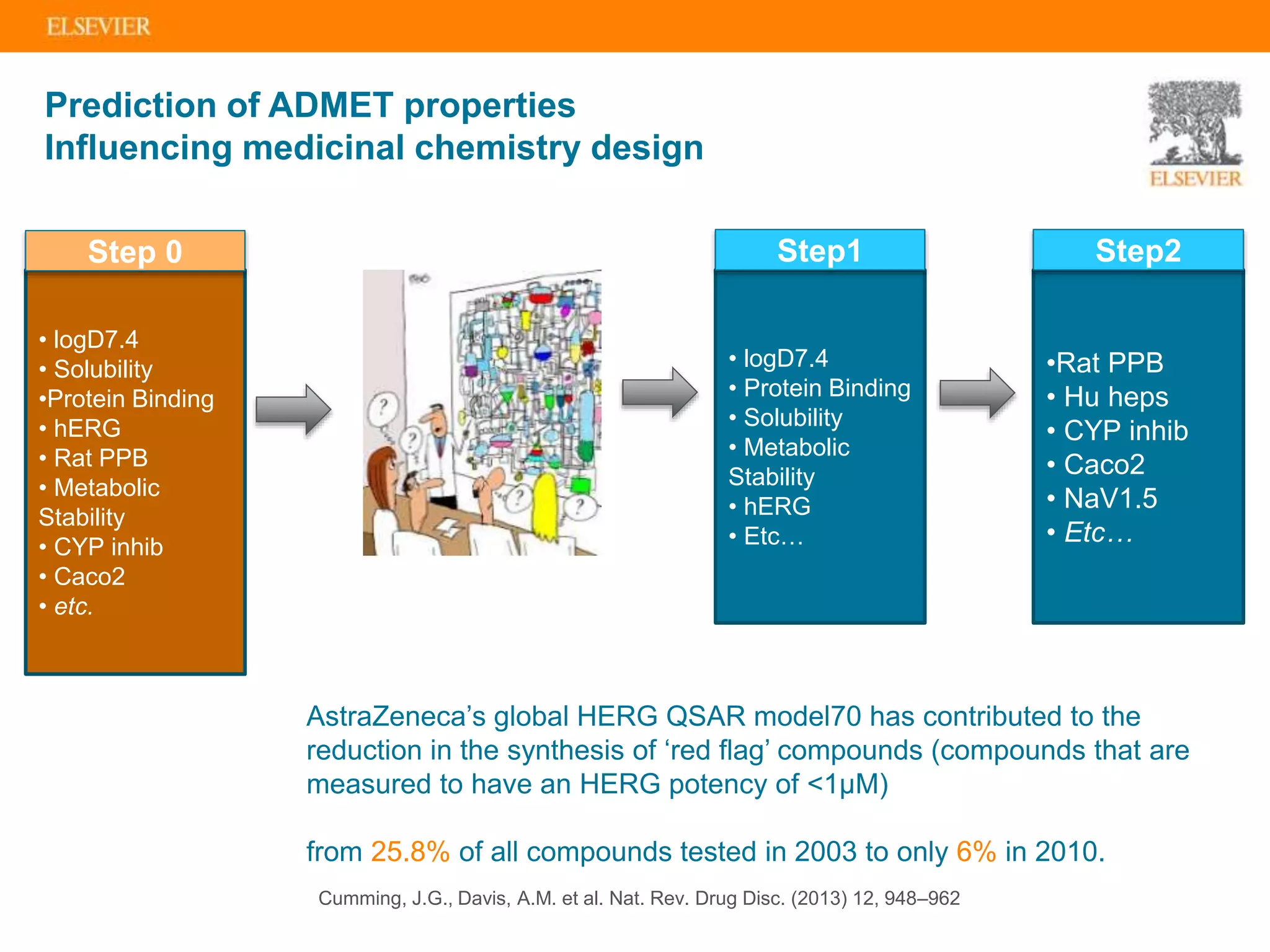
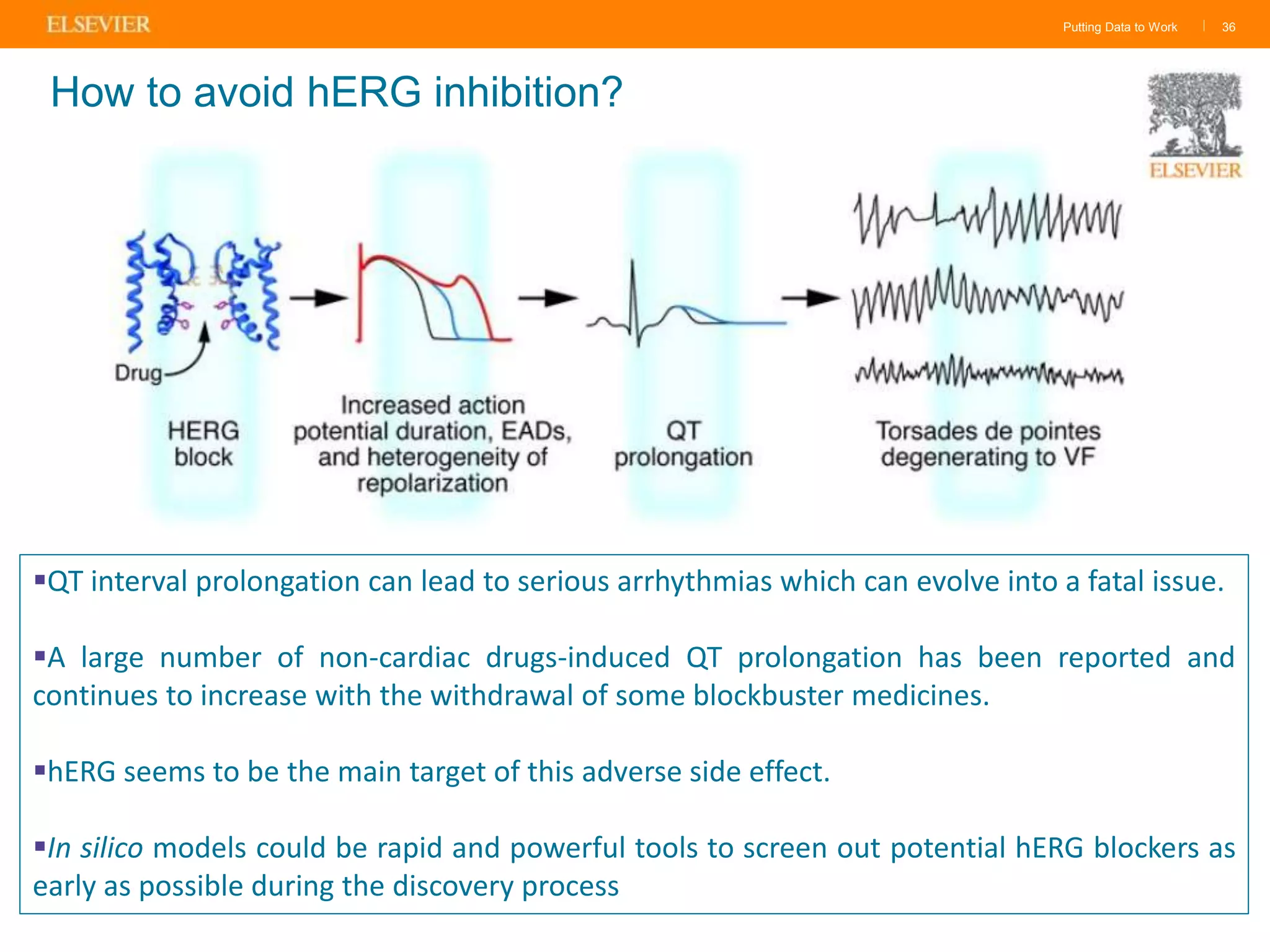
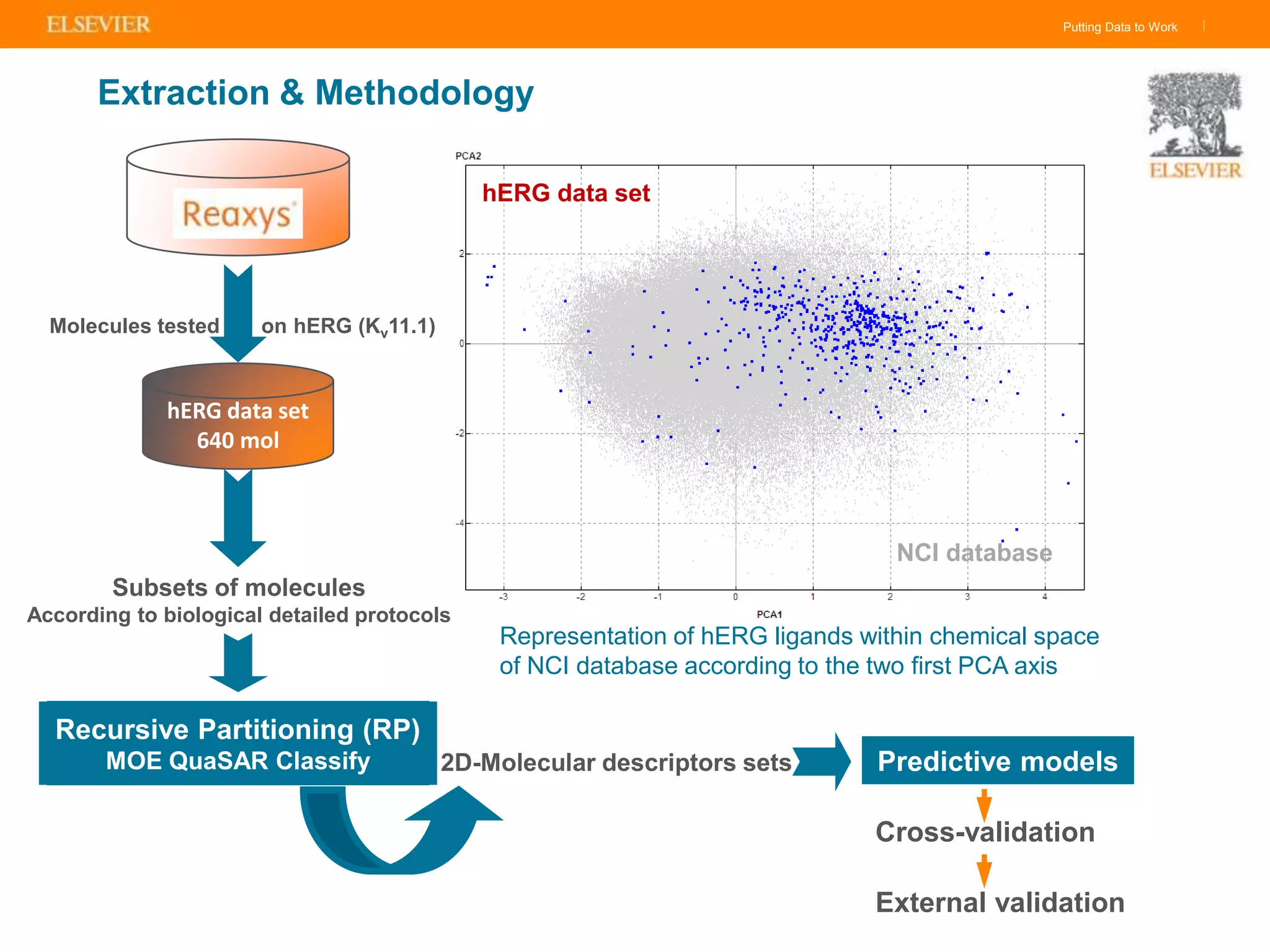
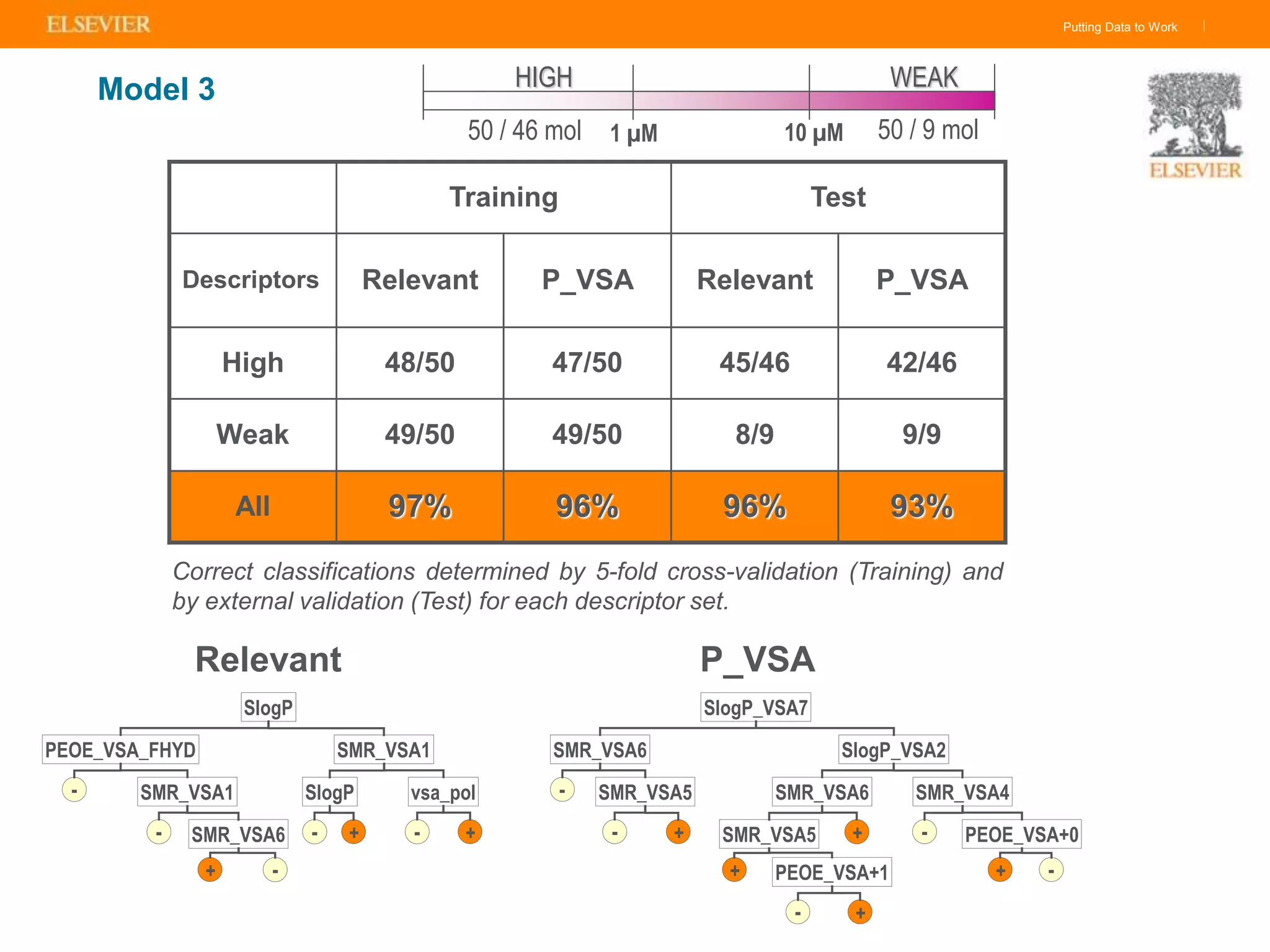
Olivier Barberan discusses data-driven drug discovery, emphasizing the importance of predictive modeling for medicinal chemists to optimize drug design and reduce attrition in pharmaceutical development. By integrating various data sources and employing virtual screening techniques, the workflow aims to enhance decision-making at all stages of drug discovery, from lead identification to clinical trials. The presentation highlights the challenges pharmaceuticals face in utilizing data effectively to improve outcomes and efficiencies in drug development.