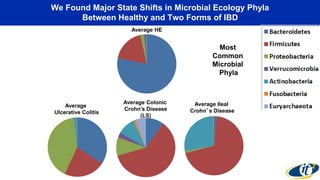
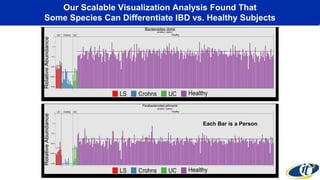
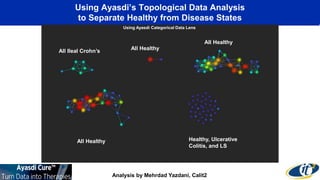
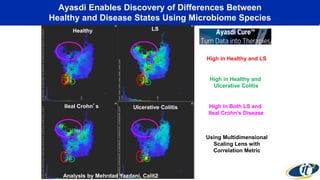
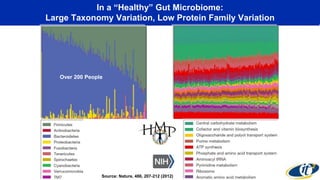
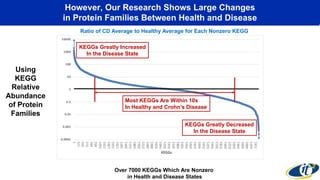
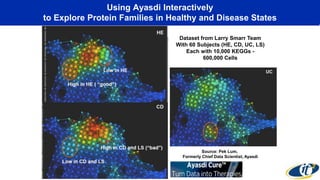
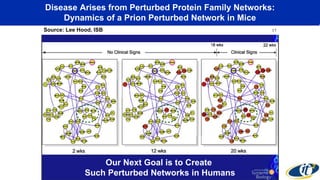
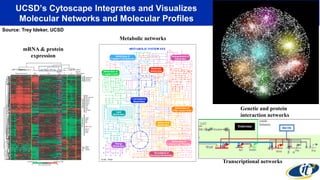
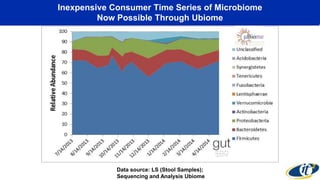
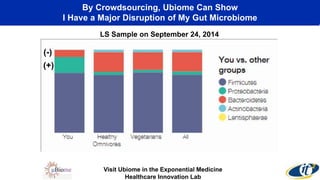
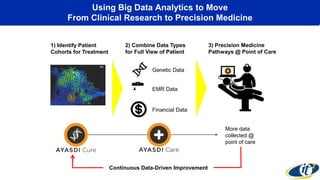
This document summarizes a talk on analyzing human microbiome data to better understand health and disease. It discusses how sequencing and supercomputing is used to map microbial ecology in hundreds of people. Advanced analytics tools like Ayasdi are helping discover patterns separating healthy from disease states like inflammatory bowel disease. Future goals include applying these techniques to larger datasets and using molecular networks to better understand disease development at the genetic and protein level.