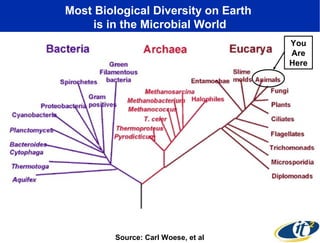
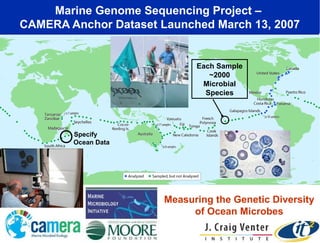
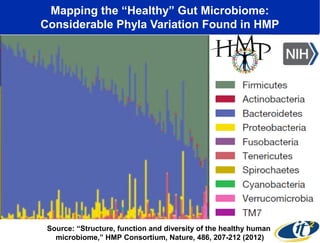
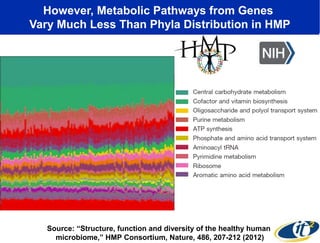
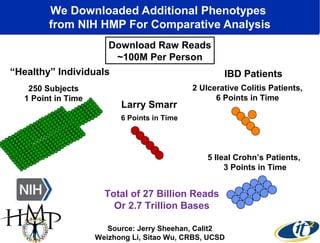
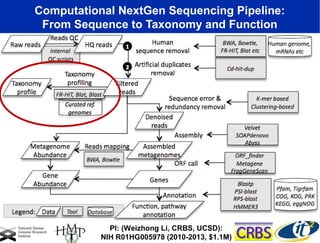
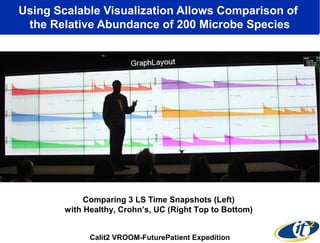
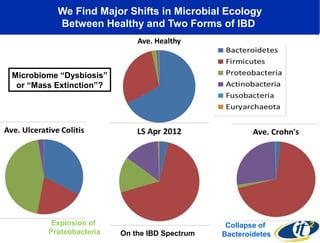
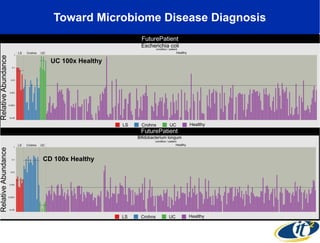
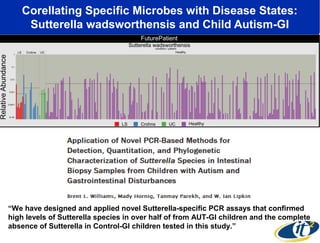
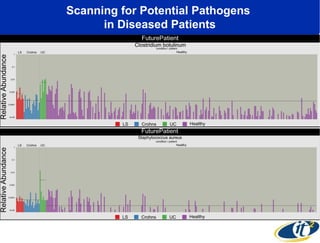
The document discusses the significance of microbial diversity in global health, highlighting the prevalence of microbes within the human body and their environmental impact. It details various research initiatives, especially in metagenomics, aimed at sequencing microbial genomes, analyzing the gut microbiome, and correlating specific microbes with diseases like Crohn's and autism. The author emphasizes the importance of data sharing and advanced computational methods for understanding microbial communities and their health implications.