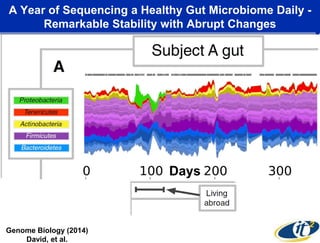
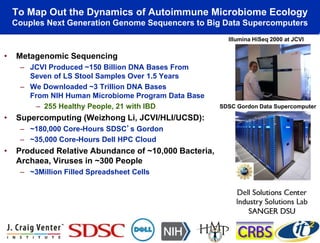
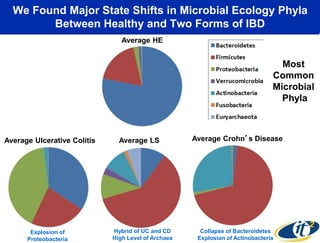
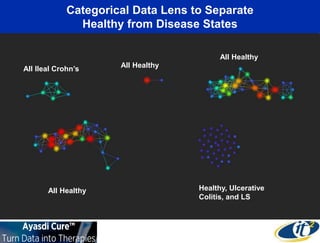
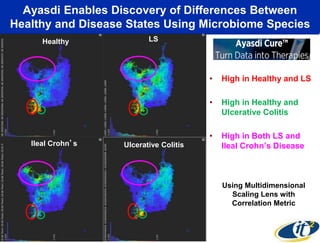
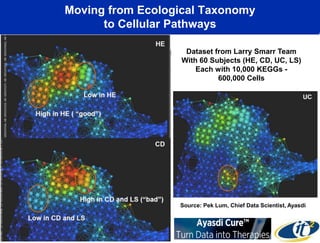
The talk by Mehrdad Yazdani discusses the revolutionary impact of mapping the human gut microbiome on medicine, emphasizing the significant presence of microbial cells and their DNA in human health. It highlights the use of cutting-edge sequencing and supercomputing technologies to analyze gut microbiome data from healthy individuals and those with inflammatory bowel disease (IBD), revealing distinct microbial state shifts. The presentation also outlines future plans to study larger microbiome datasets to identify further differences between health and disease states.