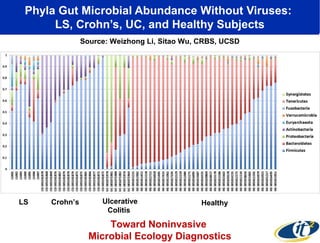
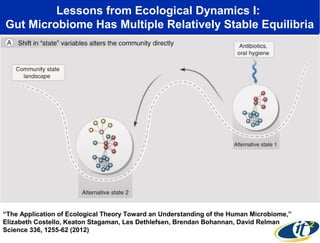
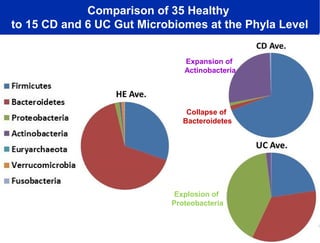
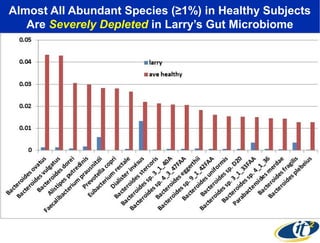
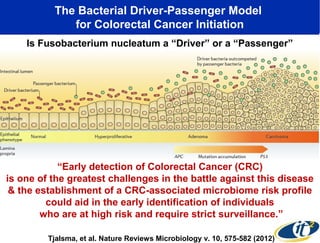
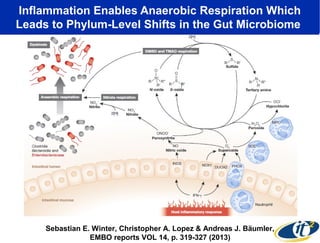
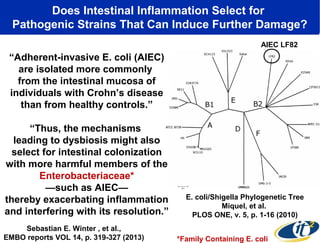
1) The document discusses a lecture given by Dr. Larry Smarr on quantifying his own "superorganism" body using big data and supercomputing.
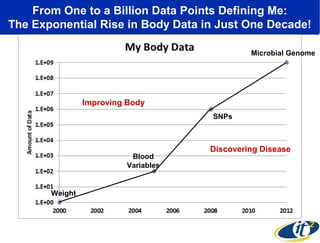
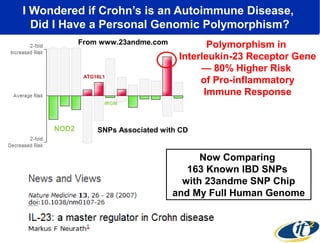
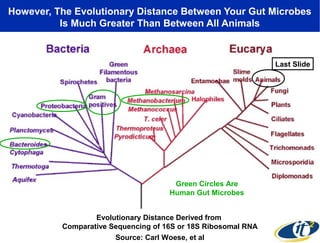
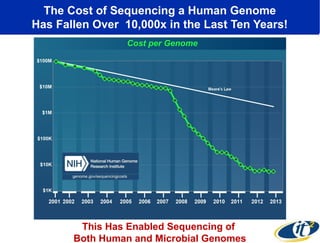
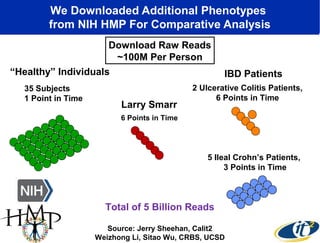
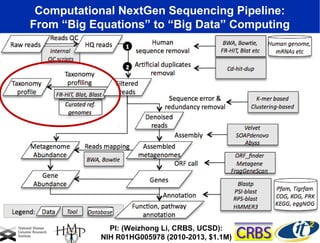
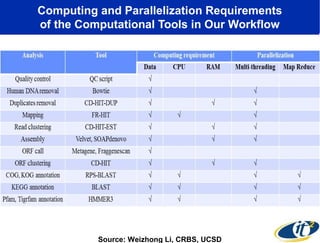
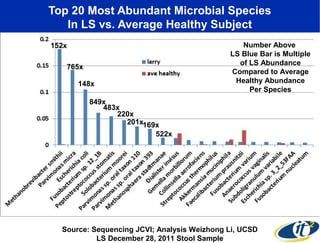
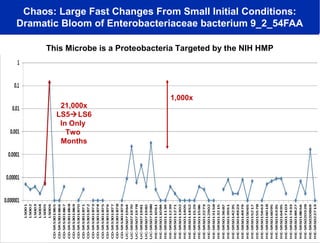
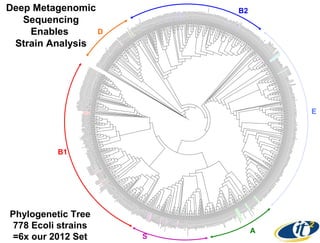
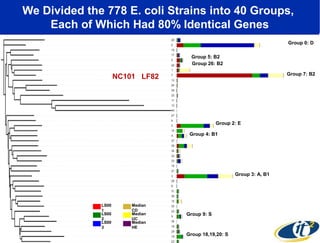
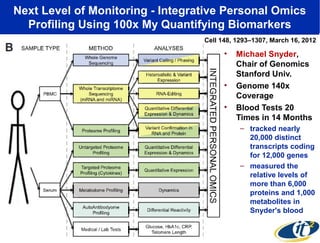
2) Over many years, Smarr collected massive amounts of biological and medical data on himself, including microbial genome sequencing of stool samples.
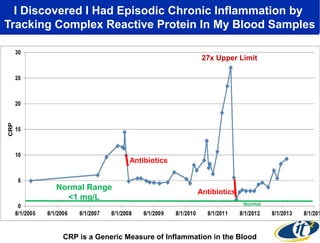
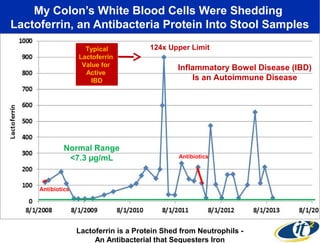
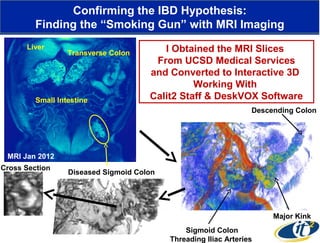
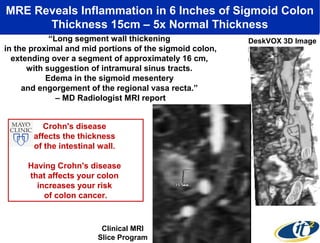
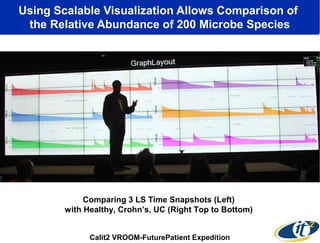
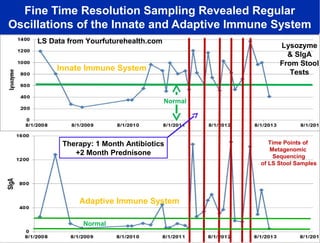
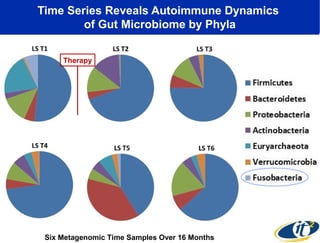
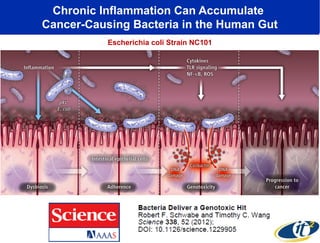
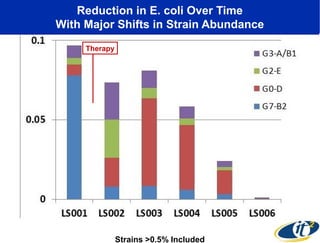
3) Analyzing this personal data using supercomputers revealed Smarr had an undiagnosed autoimmune disease (inflammatory bowel disease), disruptions to his gut microbiome, and periodic inflammation.