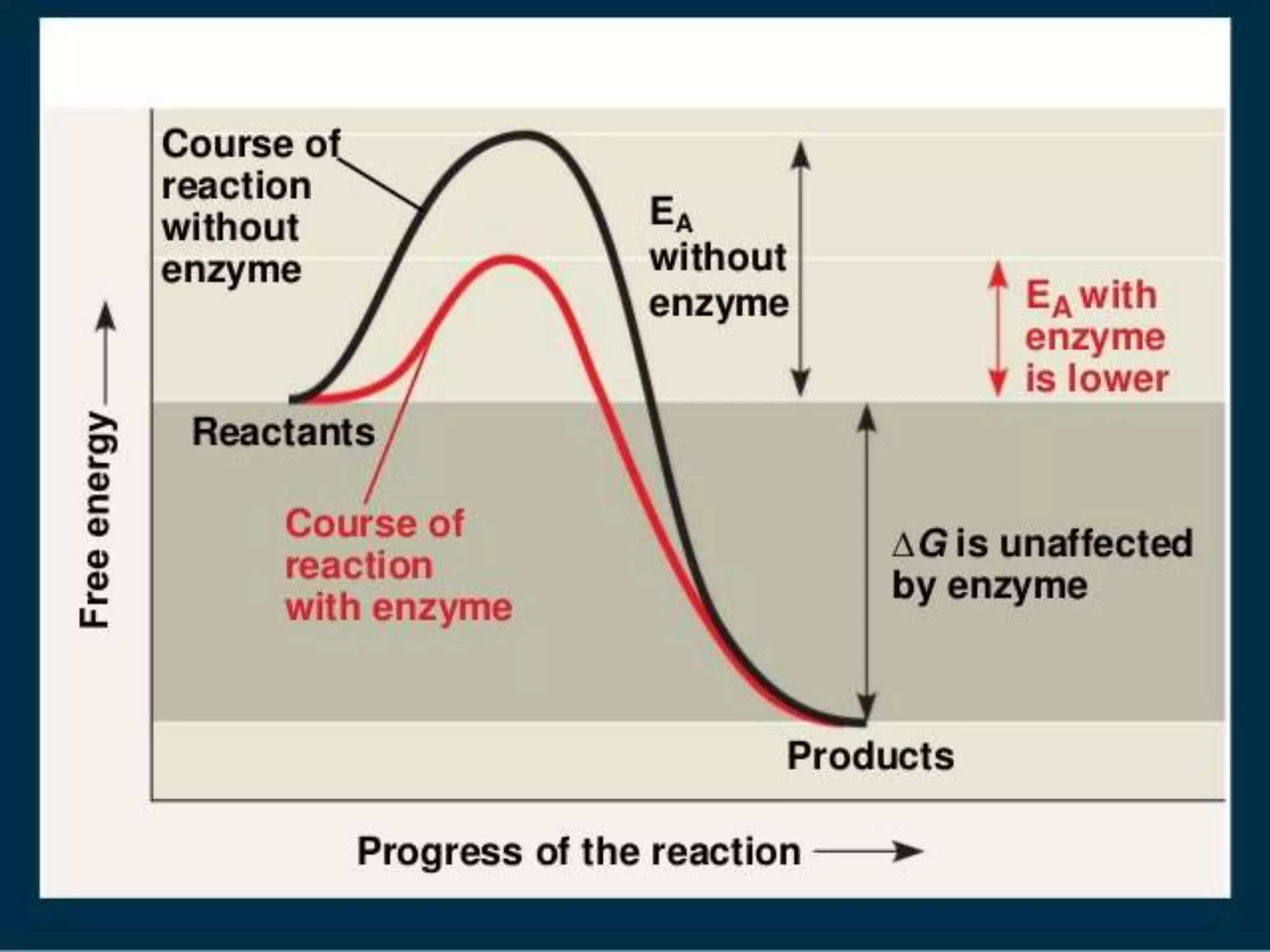
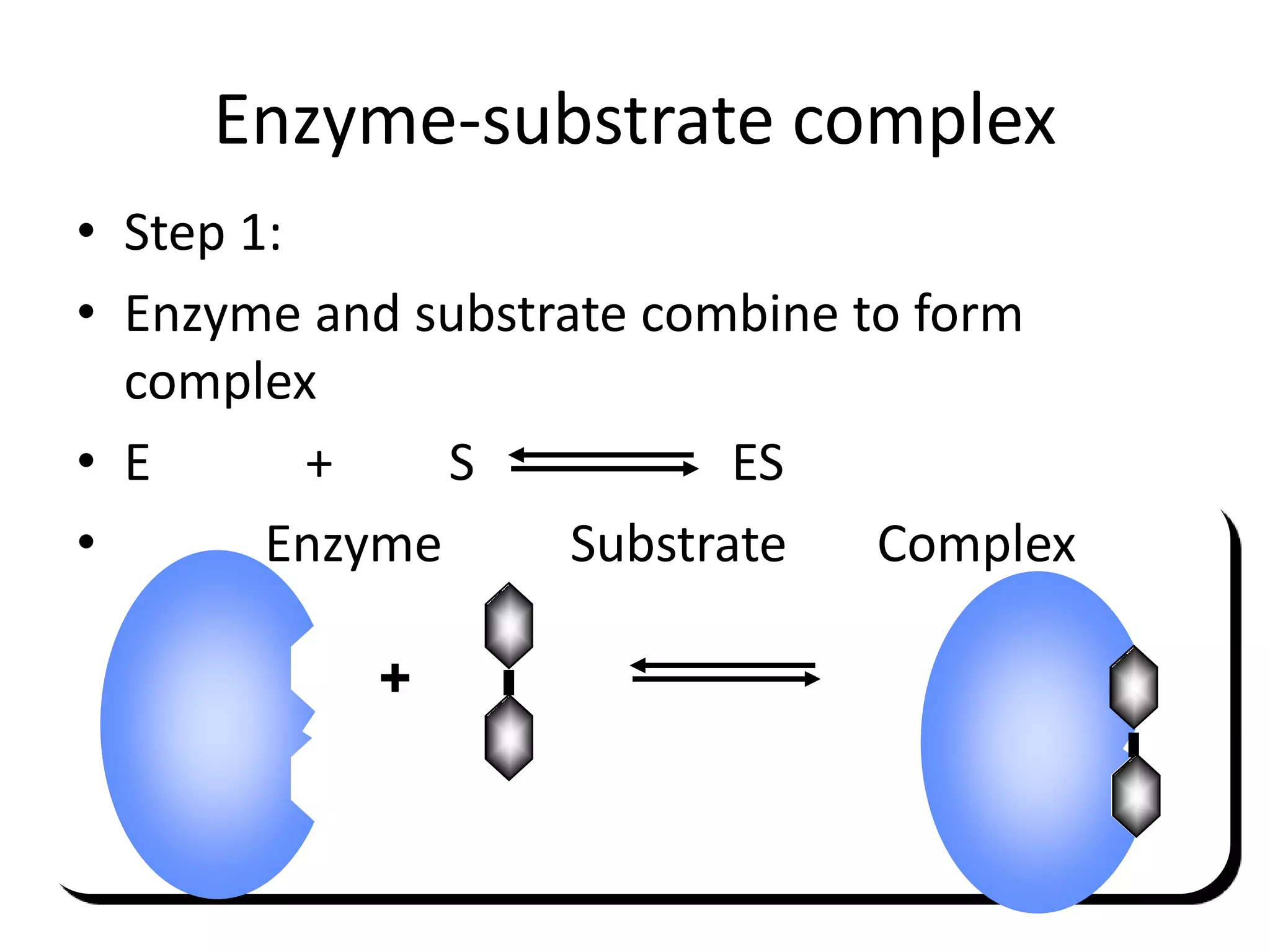
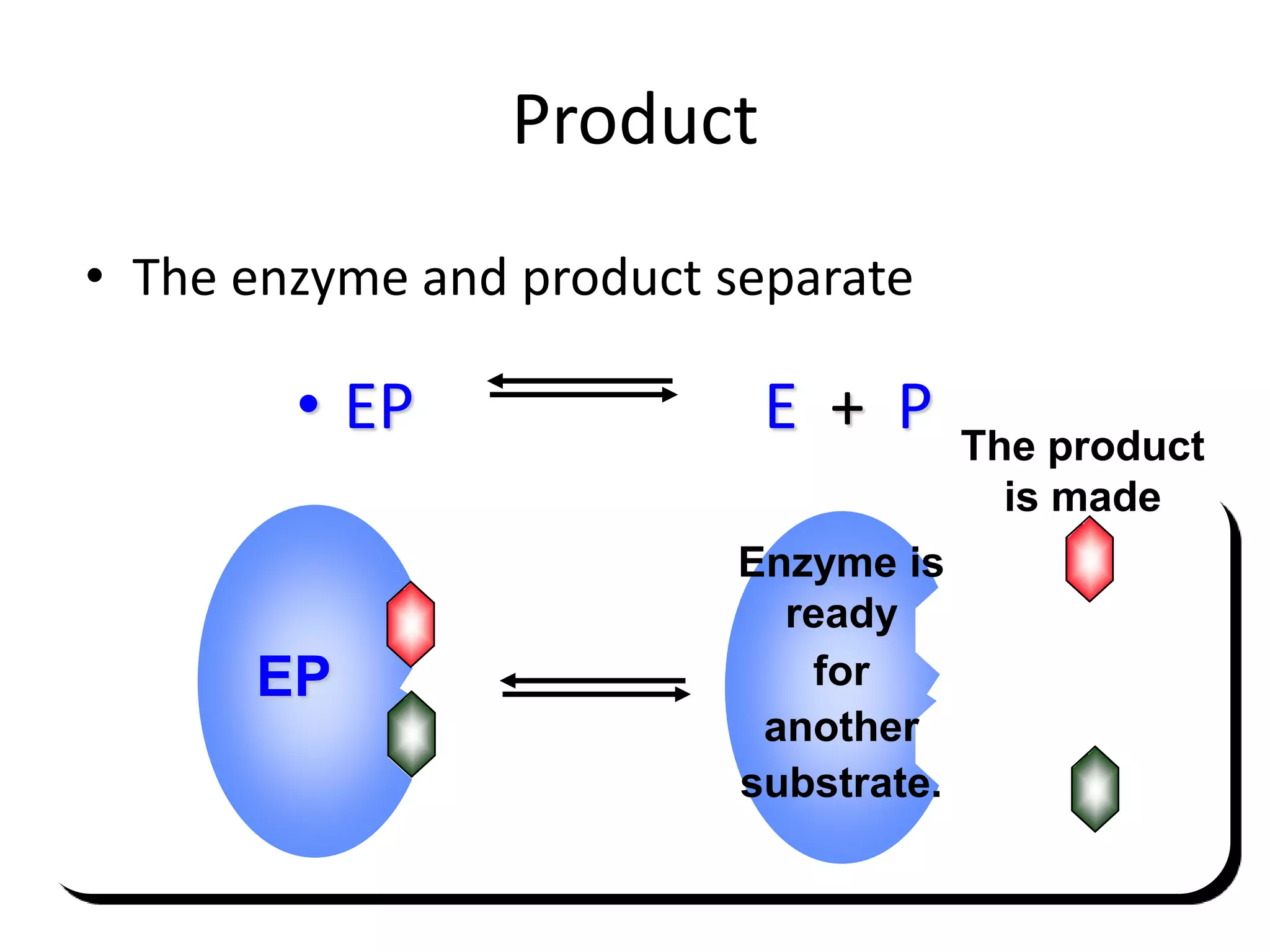
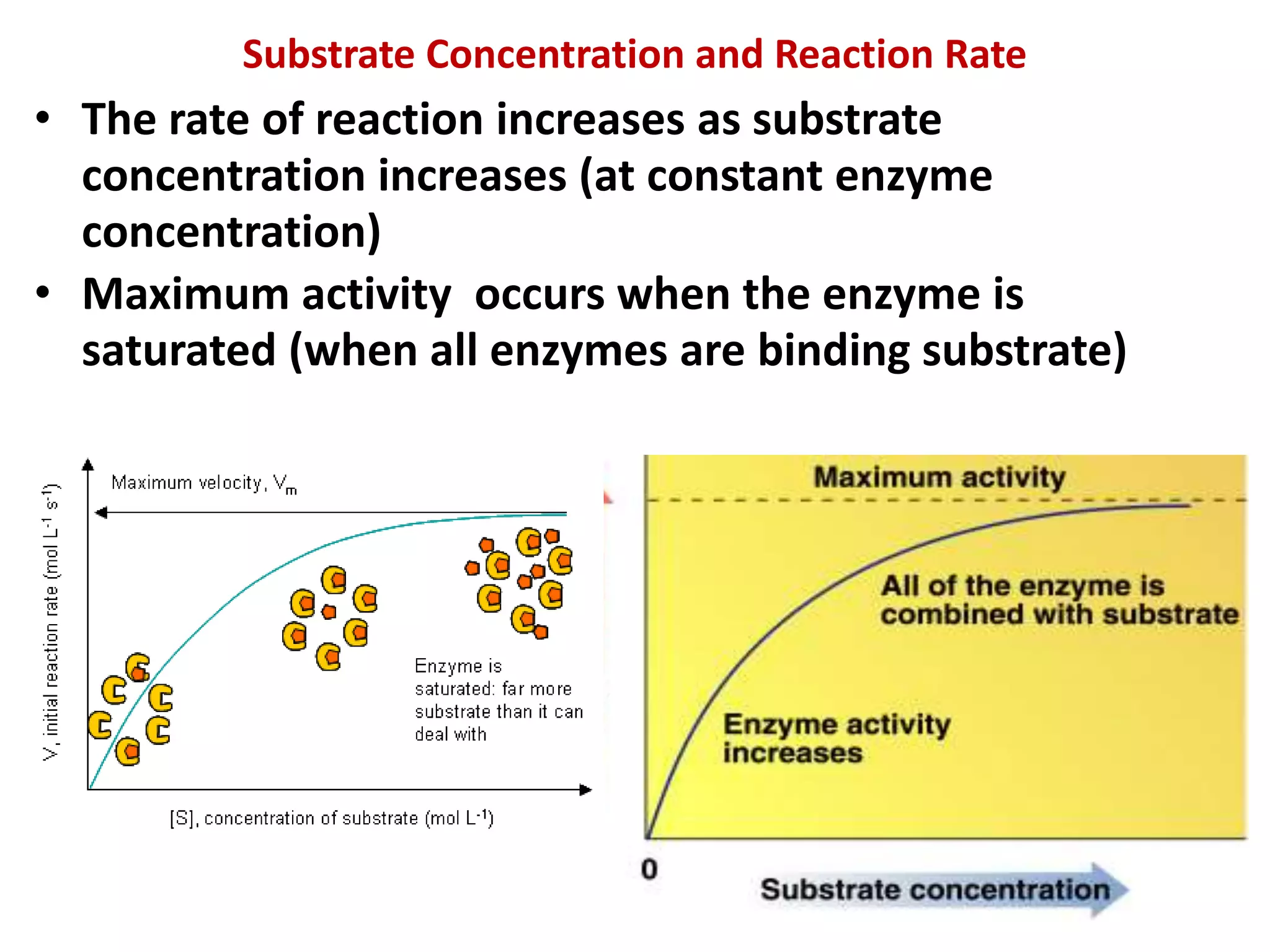
1. Enzymes are protein catalysts that lower the activation energy of biochemical reactions and increase their rates without being consumed.
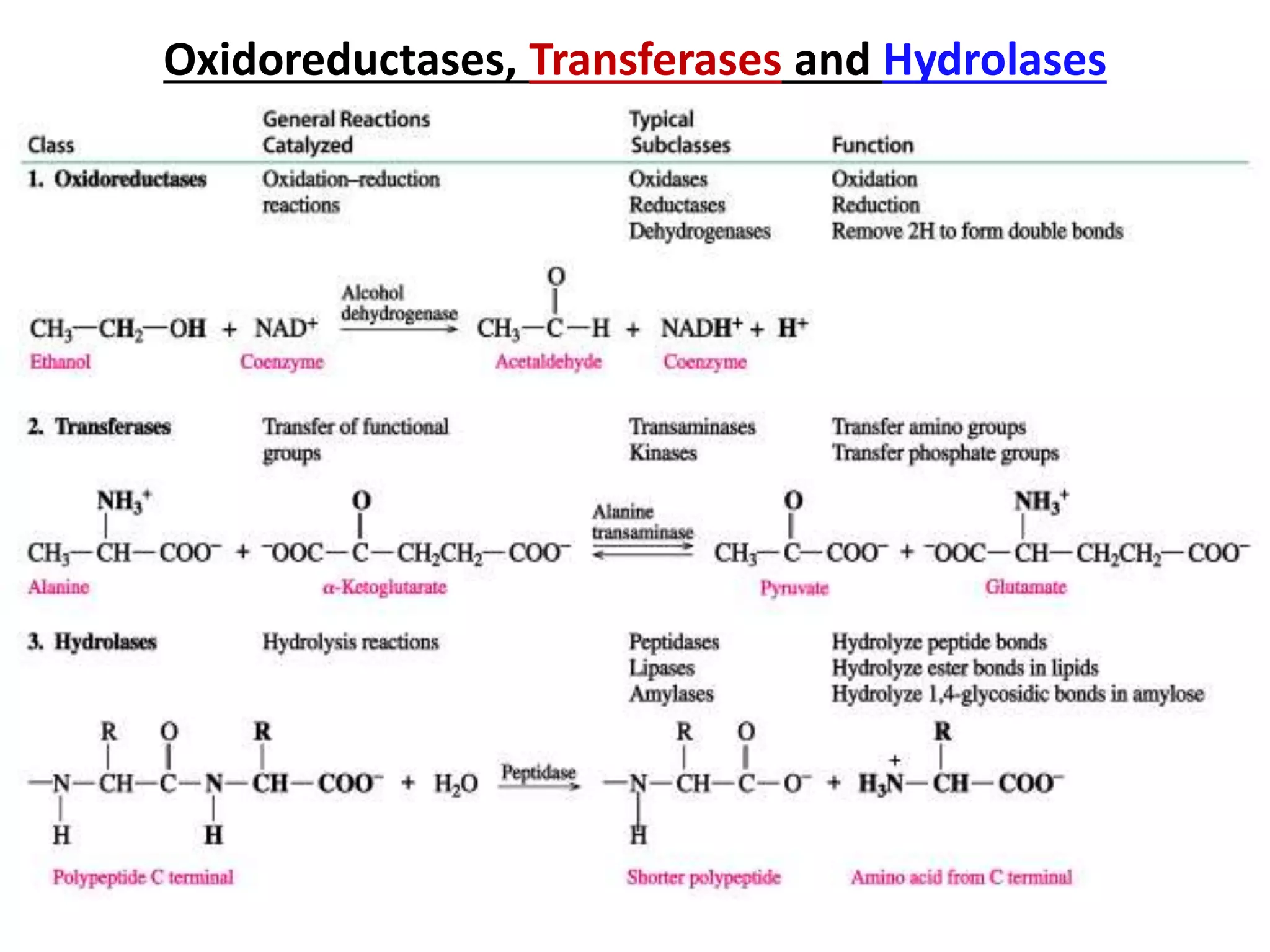
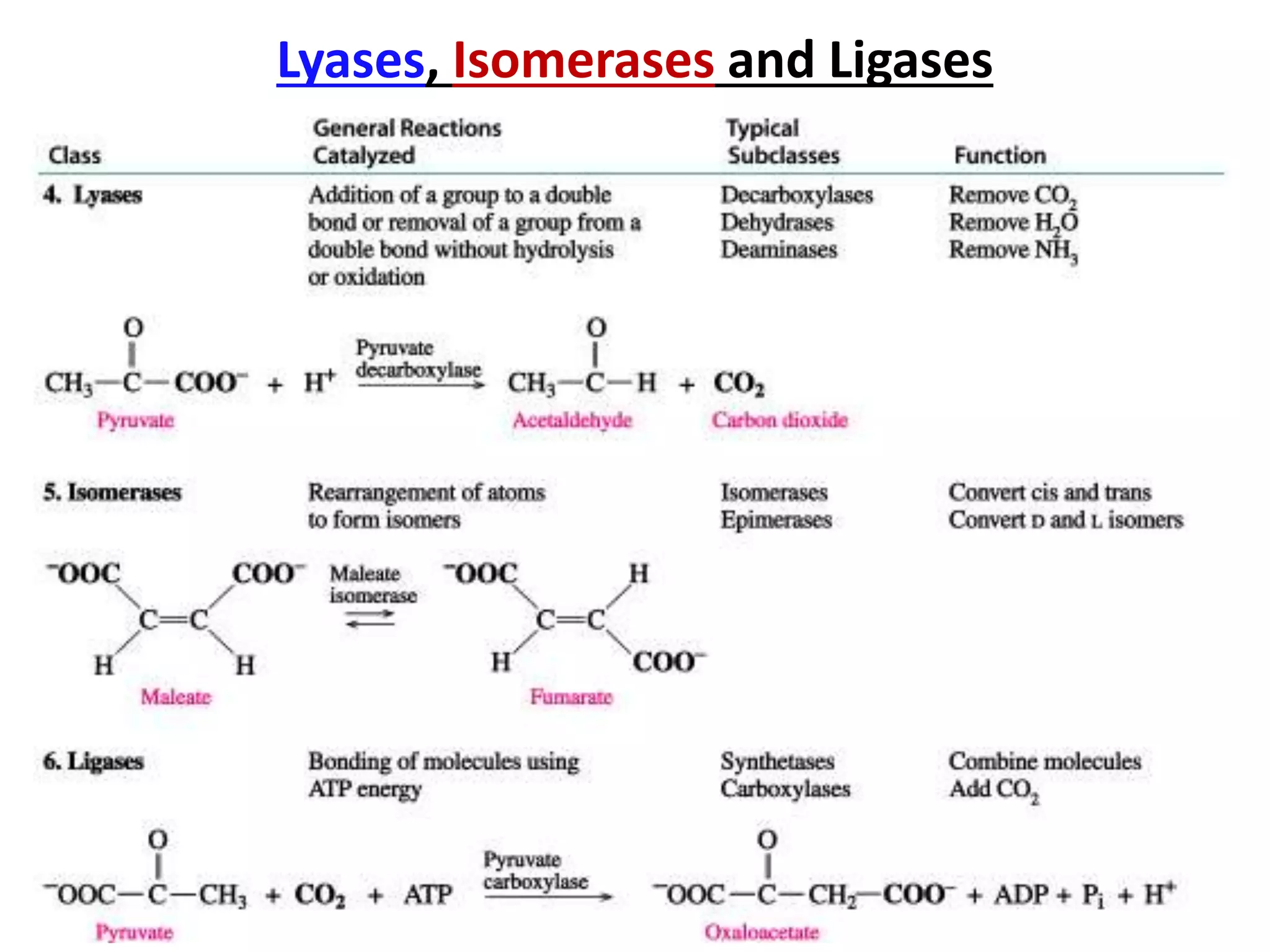
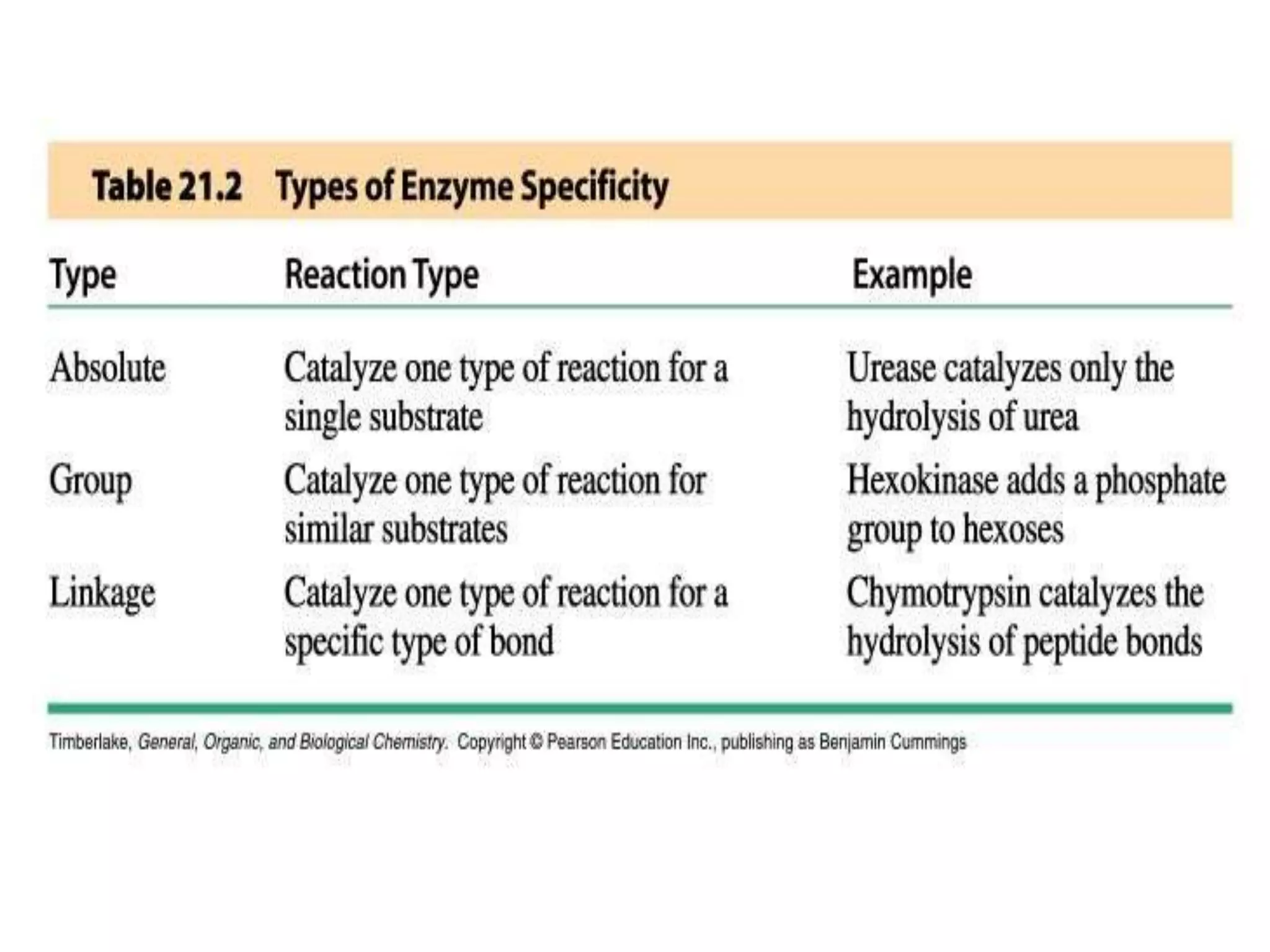
2. They are classified into six main categories based on the type of reaction they catalyze.
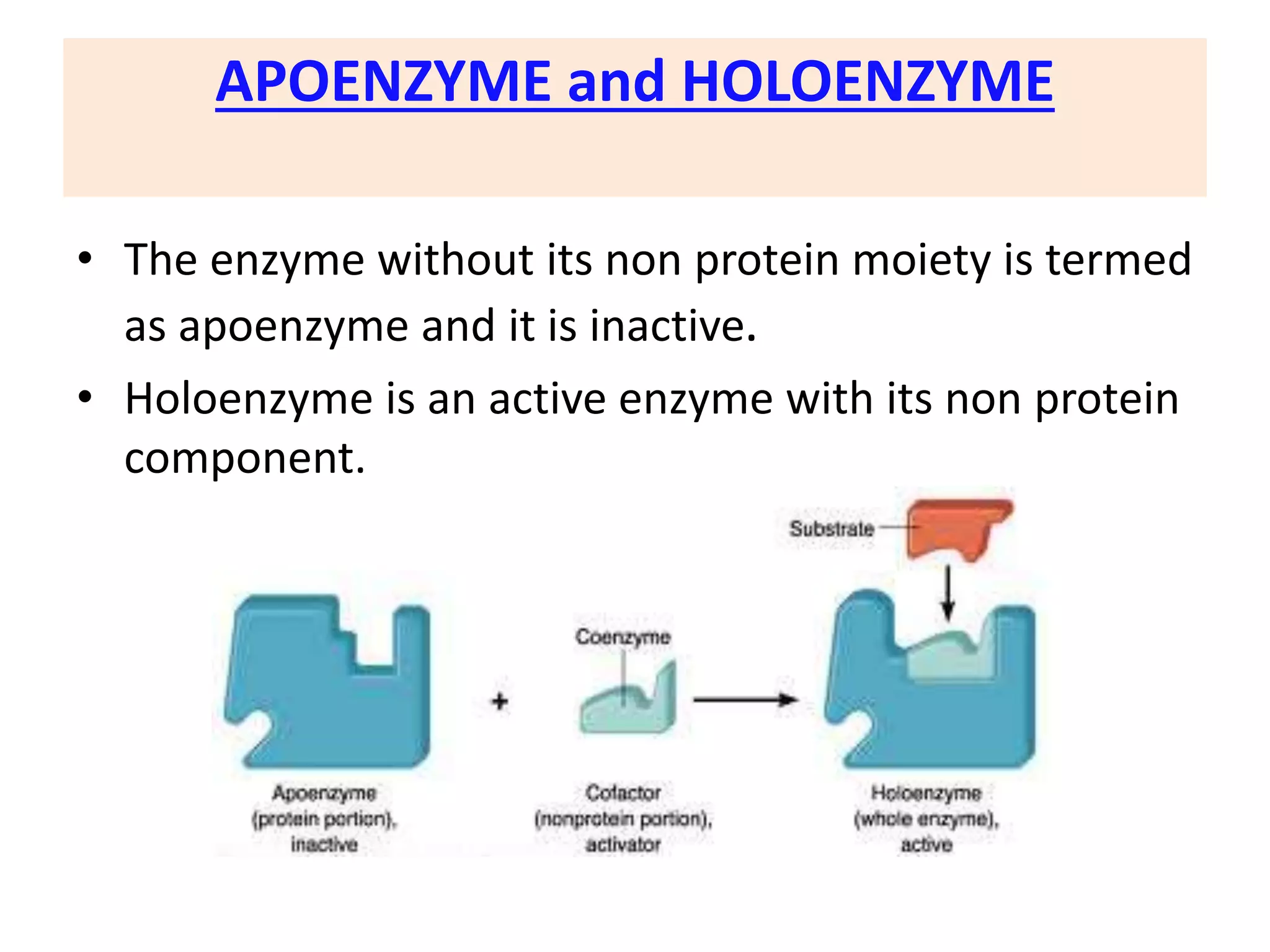
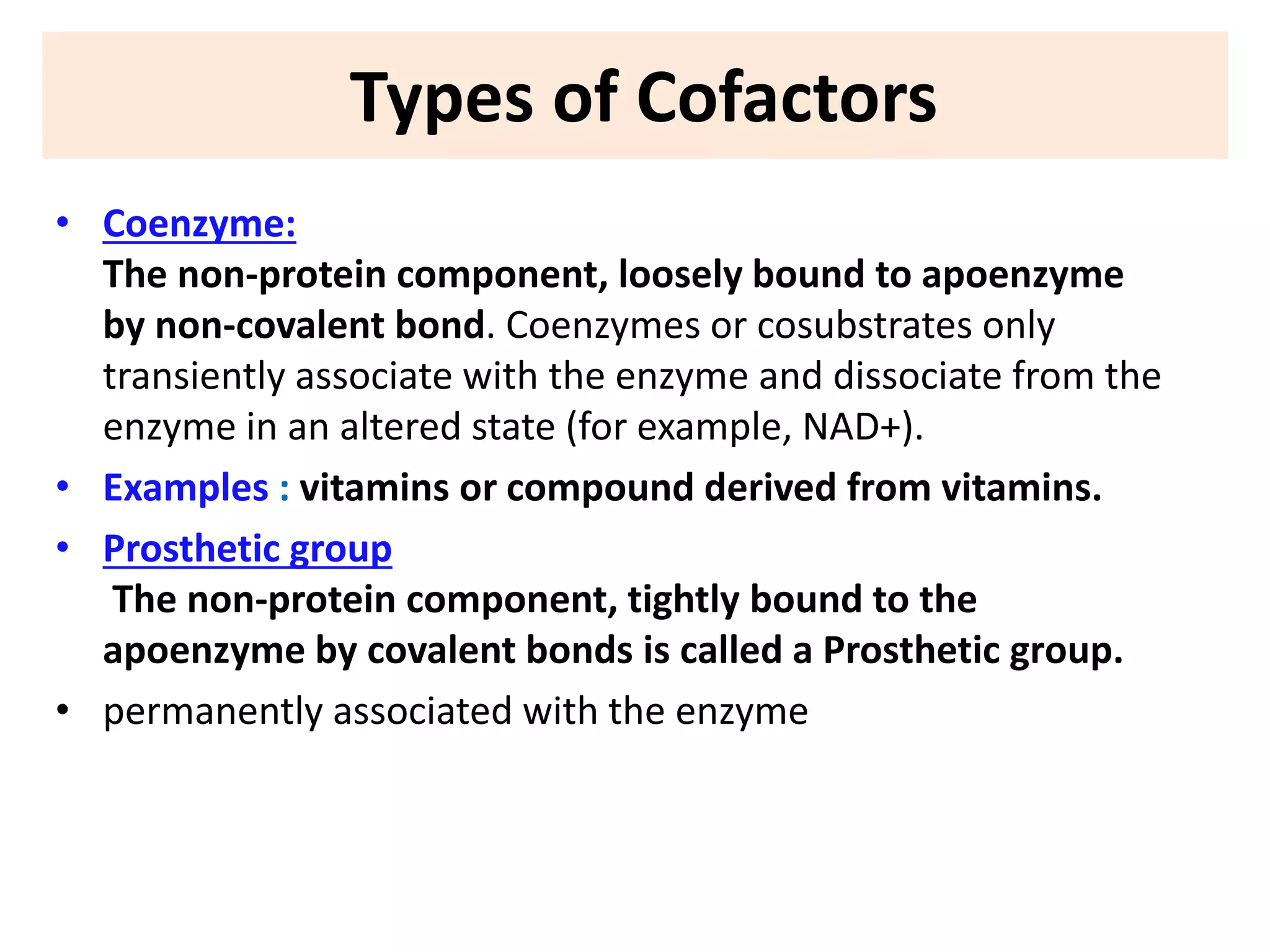
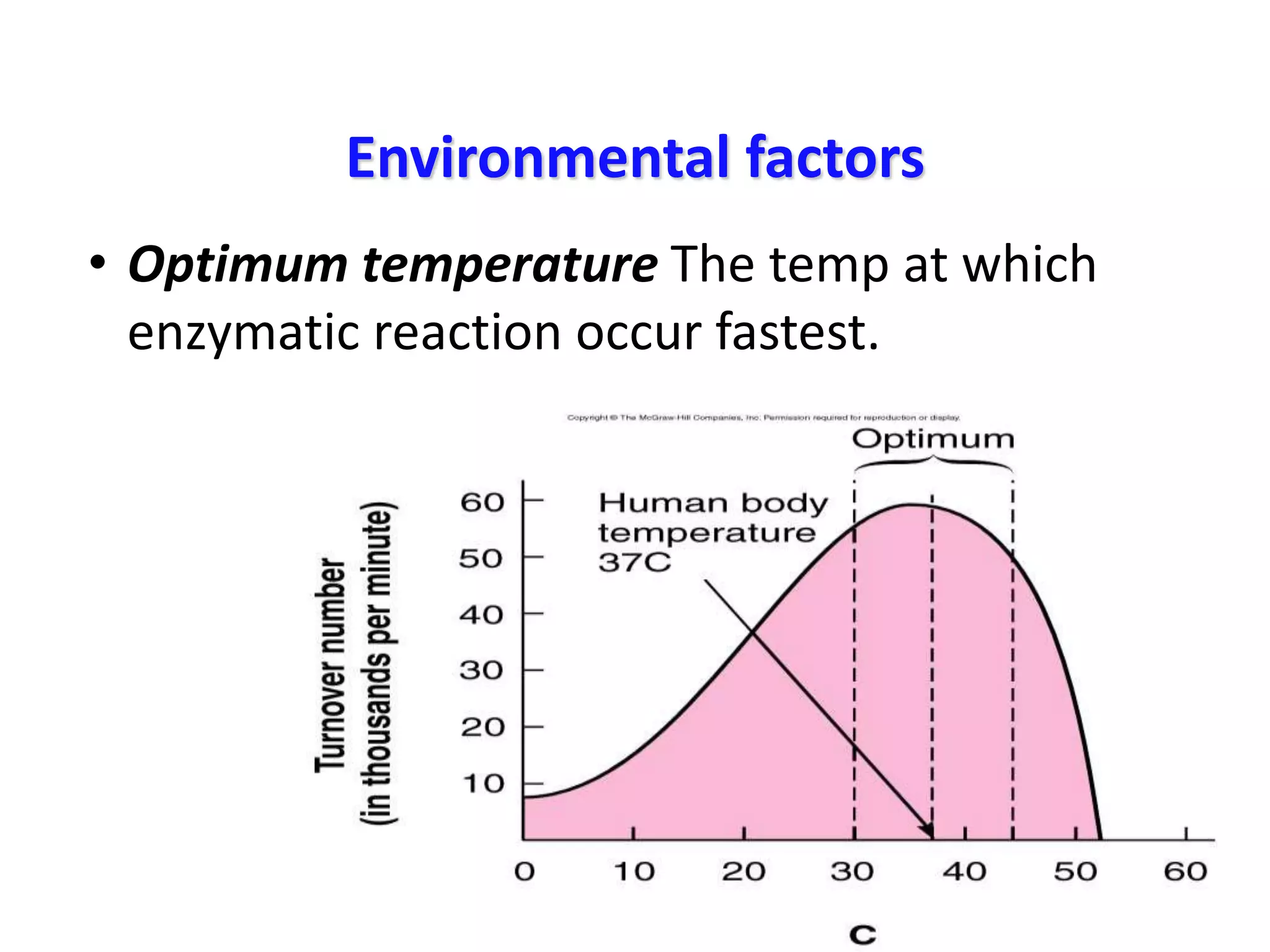
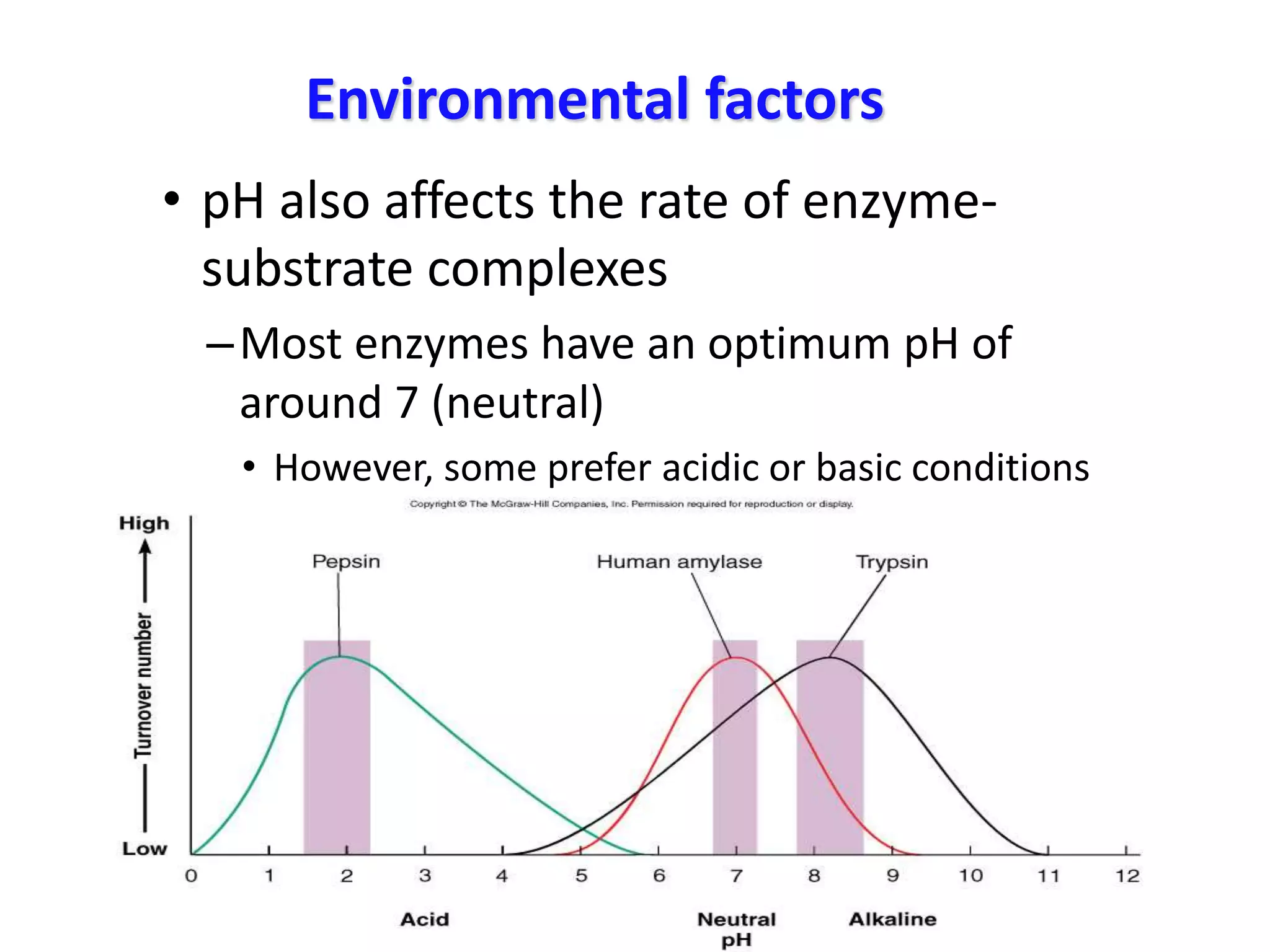
3. Enzyme activity is affected by environmental factors like temperature and pH, as well as cofactors that bind enzymes. Inhibitors can also decrease enzyme activity.


















![Michaelis-Menten Kinetics
V
[S]
V = Reaction velocity
Rate of P formation
Vmax
V = Vm* [S]
Km + [S]](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-31-2048.jpg)



![Michaelis Constant (Km)
V = Vm * [S]
Km + [S]
Key Points:
1.Km has same units as [S]
2.At some point on graph, Km must equal [S]](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-35-2048.jpg)
![V = Vm * [S] = Vm * [S] = Vm
[S] + [S] 2 [S] 2
When V = Vm/2
[S] = Km](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-36-2048.jpg)
![Vmax
V = Vm* [S]
Km + [S]
Km
[S]](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-37-2048.jpg)
![V = Vm* [S]
Km + [S]
• Small Km Vm reached at low concentration [S]
• Large Km Vm reached at high concentration [S]
Vmax
Vmax/2
Km
[S]](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-38-2048.jpg)
![V = Vm* [S]
Km + [S]
Michaelis Constant (Km)
• Small Km Substrate binds easily at low [S]
• High affinity substrate for enzyme
• Large Km Low affinity substrate for enzyme
Vmax
Vmax/2](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-39-2048.jpg)
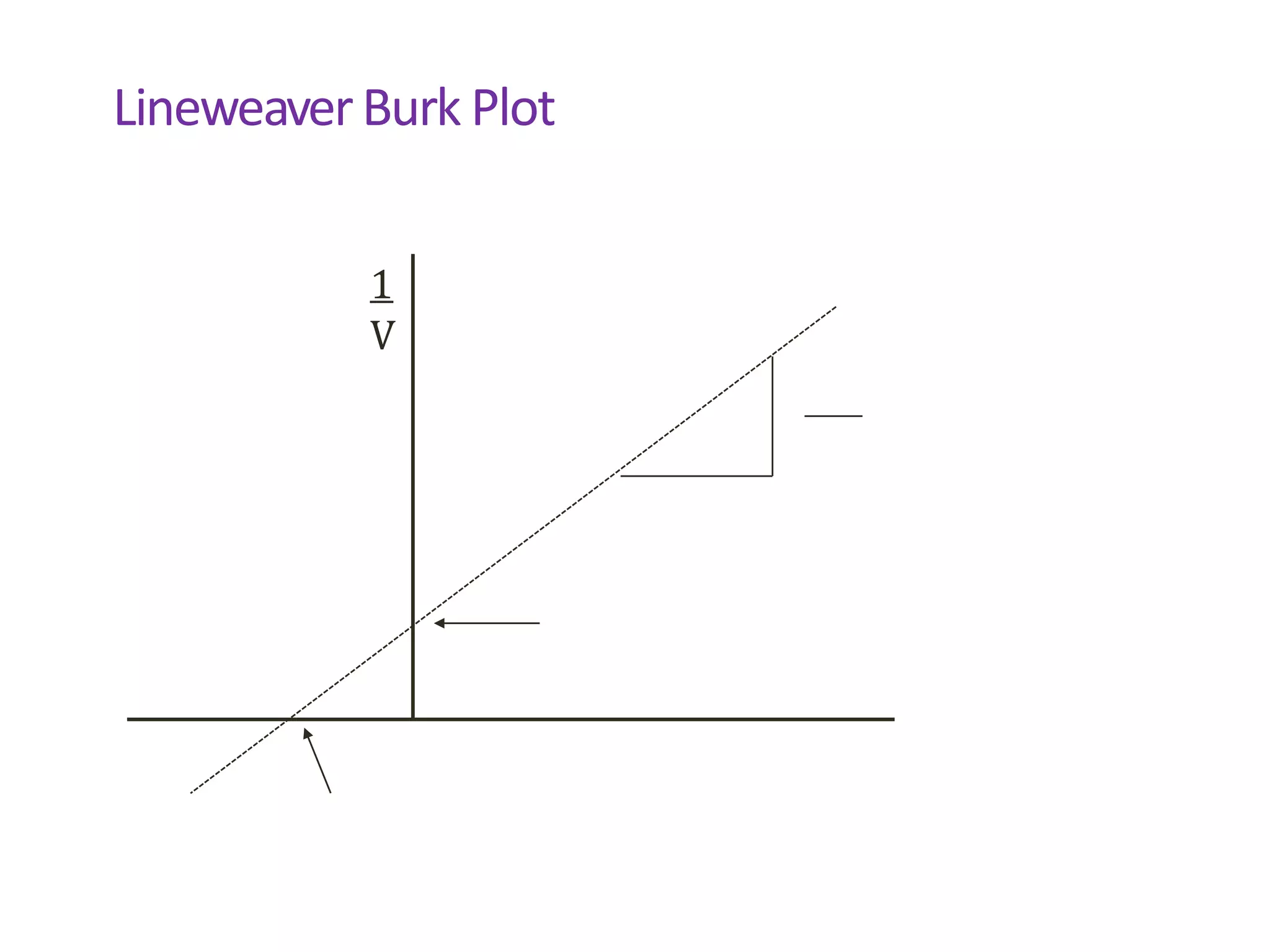
![Key Points
• Km is characteristic of each substrate/enzyme
• Vm depends on amount of enzyme present
• Can determine Vm/Km from
• Michaelis Menten plot V vs. [S]
• Lineweaver Burk plot 1/V vs. 1/[S]](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-40-2048.jpg)
![Lineweaver Burk Plot
V = Vm* [S]
Km + [S]
1 = Km + [S] =
V Vm [S]
Km + [S]
Vm [S] Vm[S]
1 = C * 1 + 1
V [S] Vm](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-41-2048.jpg)
![Competitive Inhibitor
[S]
Vmax
Vmax/2
Km
Normal
Km
Same Vm
Higher Km
Inhibitor](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-44-2048.jpg)
![Vmax
With inhibitor
Vmax
Vmax/2
Vmax/2
Lower Vm
Same Km
Km
[S]
Non-competitive Inhibitor](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-45-2048.jpg)


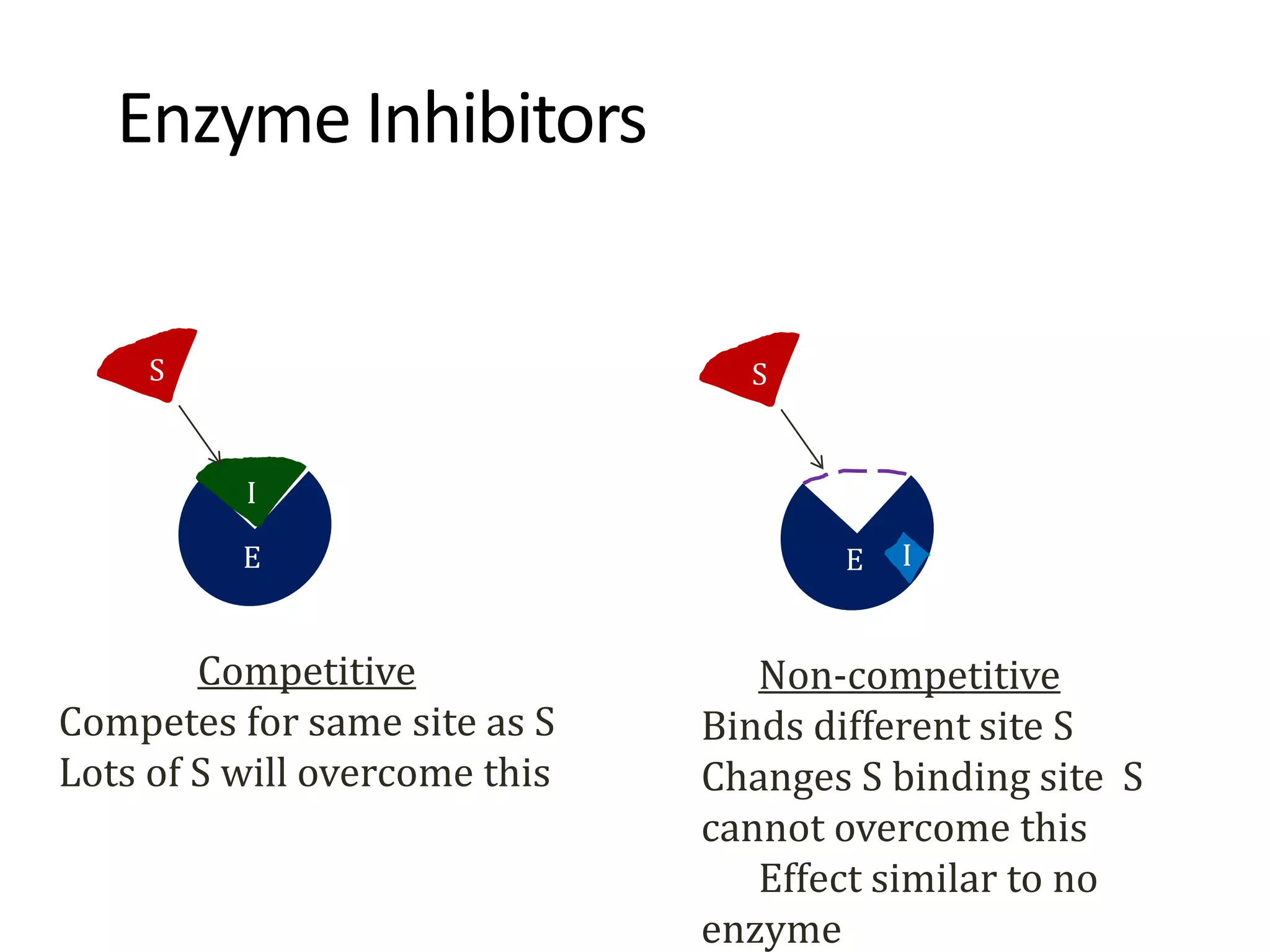
![Competitive inhibition
• This type of inhibition occurs when the inhibitor binds reversibly to the
same site that the substrate would normally occupy and, therefore,
competes with the substrate for binding to the enzyme active site.
• 1. Effect on Vmax: The effect of a competitive inhibitor is reversed by
increasing the concentration of substrate. At a sufficiently high [S], the
reaction velocity reaches the Vmax observed in the absence of inhibitor,
that is, Vmax is unchanged in the presence of a competitive inhibitor
• 2. Effect on Km: A competitive inhibitor increases the apparent Km for a
given substrate. This means that, in the presence of a competitive
inhibitor, more substrate is needed to achieve half Vmax.](https://image.slidesharecdn.com/enzymespres-230120184721-be254982/75/enzymes-pres-ppt-48-2048.jpg)