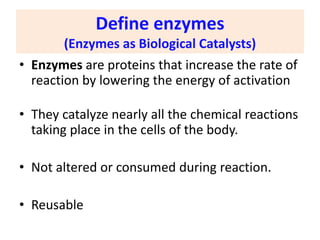
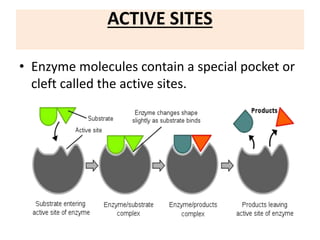
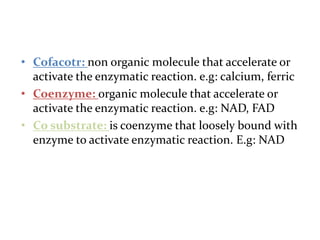
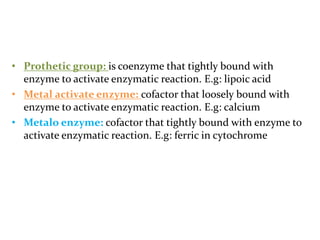
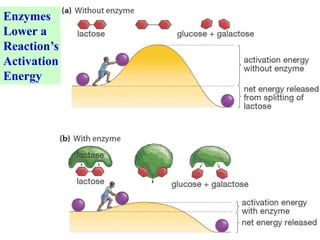
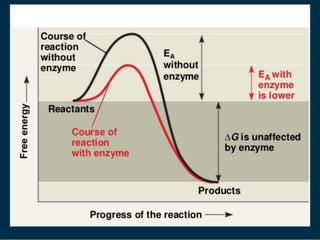
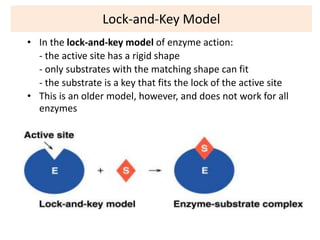
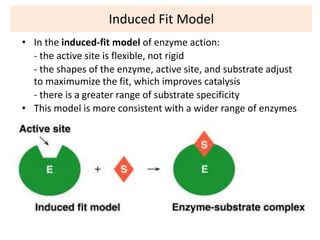
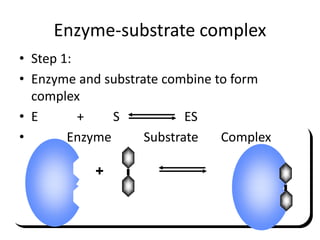
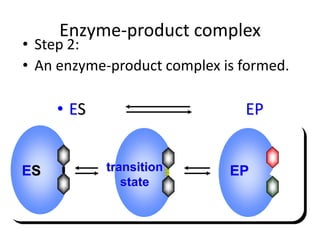
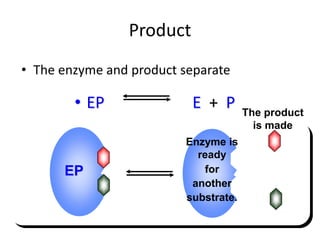
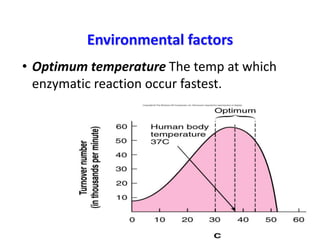
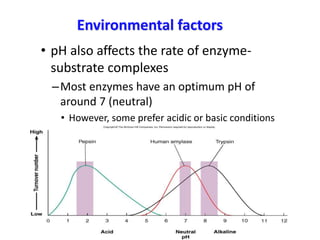
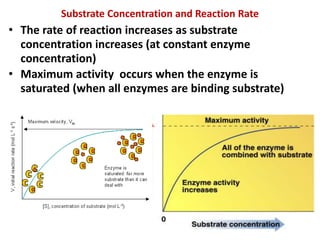
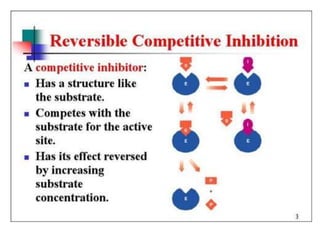
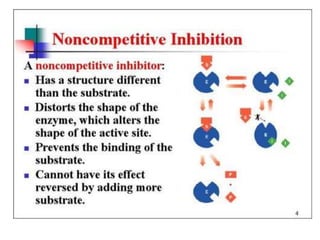
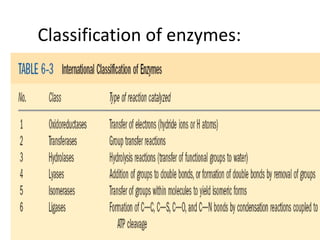
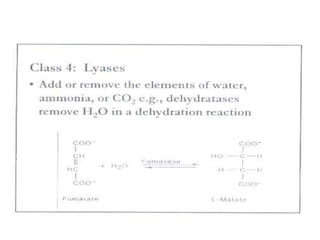
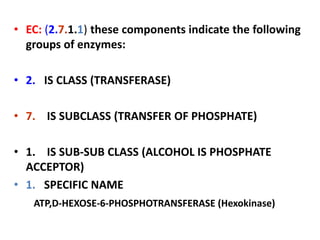
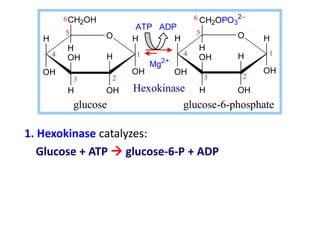
This document discusses enzymes, including their structure, classification, and mechanisms of action. It defines enzymes as proteins that lower the activation energy of chemical reactions and notes they are not consumed during reactions. It describes important enzyme components like active sites and cofactors/coenzymes. It explains environmental factors and inhibitors that can affect enzyme activity. Different classification systems are presented, and key terms like substrate specificity and enzyme naming conventions are outlined.