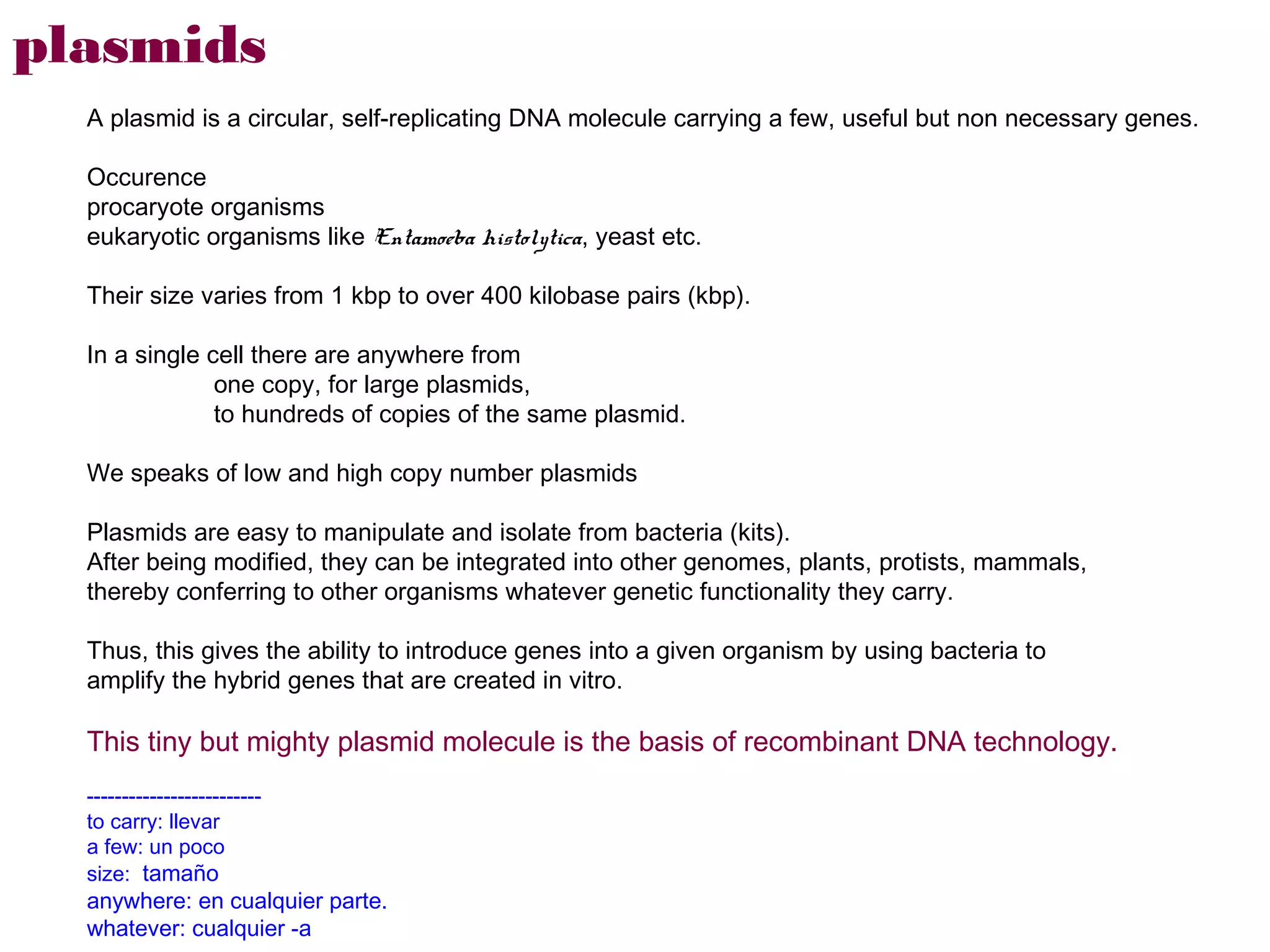
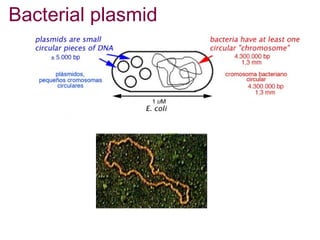
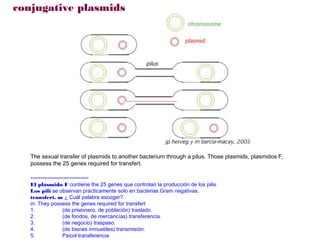
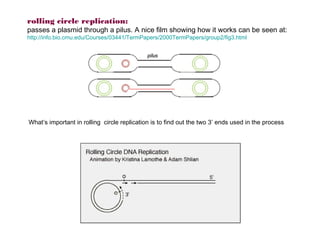
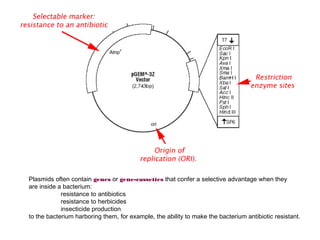
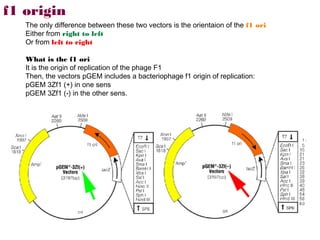
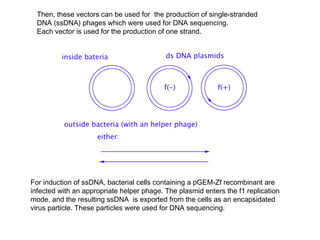
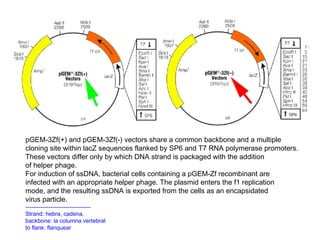
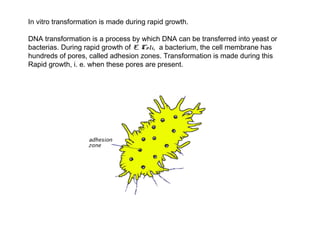
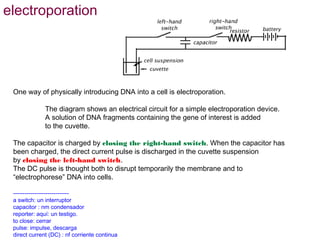
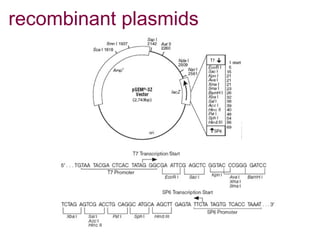
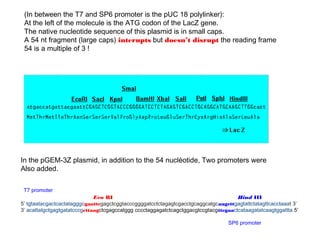
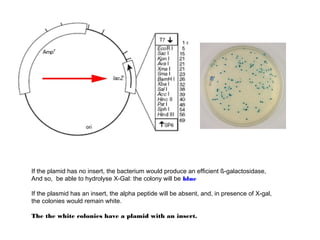
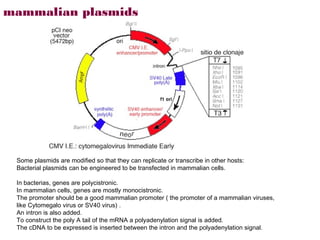
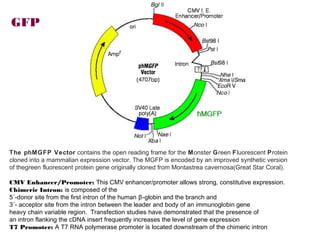
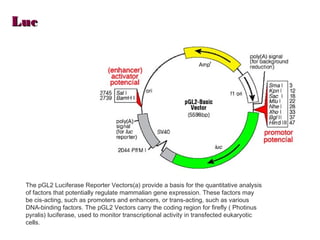
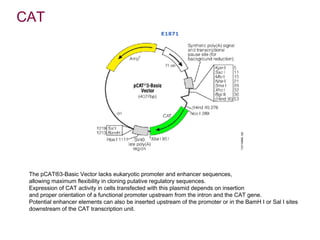
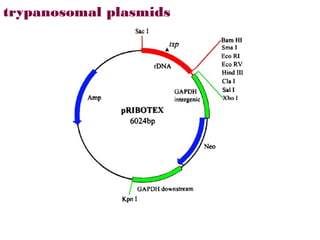
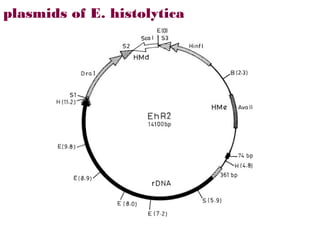
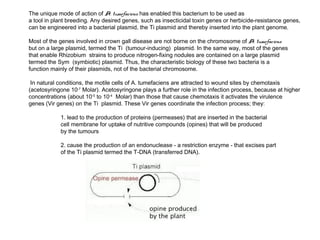
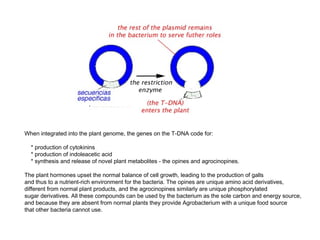
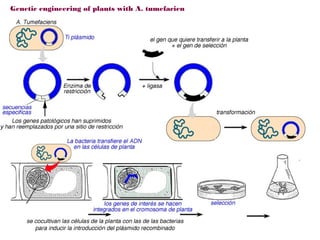
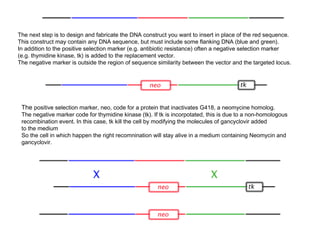
Here are the key steps to open the plasmid polylinker using restriction enzymes:
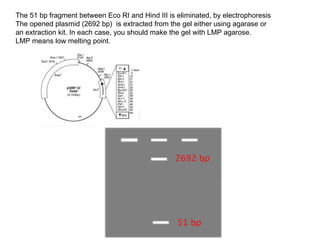
1. Digest the plasmid with EcoRI and HindIII restriction enzymes and their appropriate buffer.
2. This will cut the plasmid at the EcoRI and HindIII sites, linearizing the plasmid and removing a 51 bp fragment from the polylinker region.
3. Run the digested plasmid on an agarose gel to separate the linearized plasmid from the excised 51 bp fragment.
4. Isolate the linearized plasmid from the gel using a gel extraction kit. This prepares the plasmid with overhangs compatible for ligation of the insert.
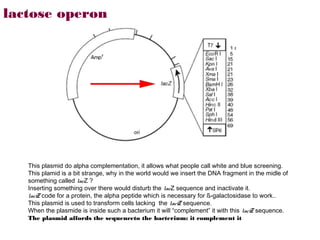
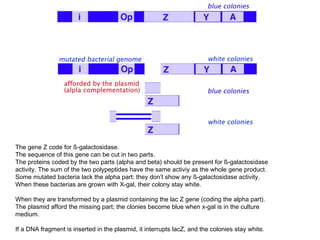
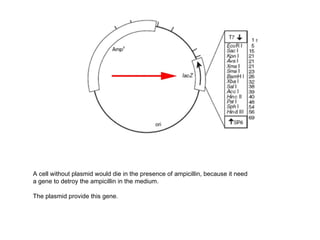
The restriction digestion opens up the polylinker region, making room for the insert DNA to