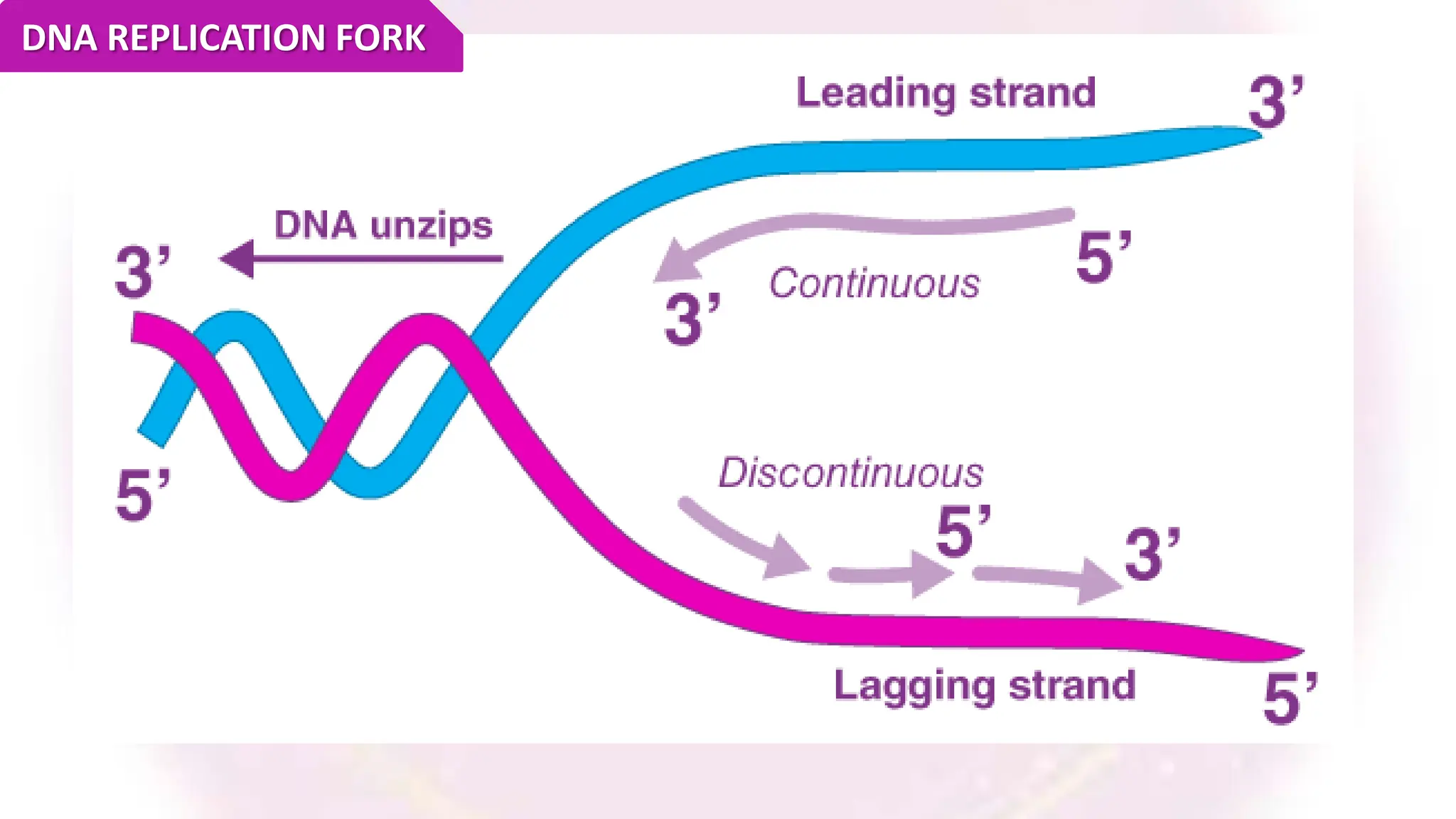
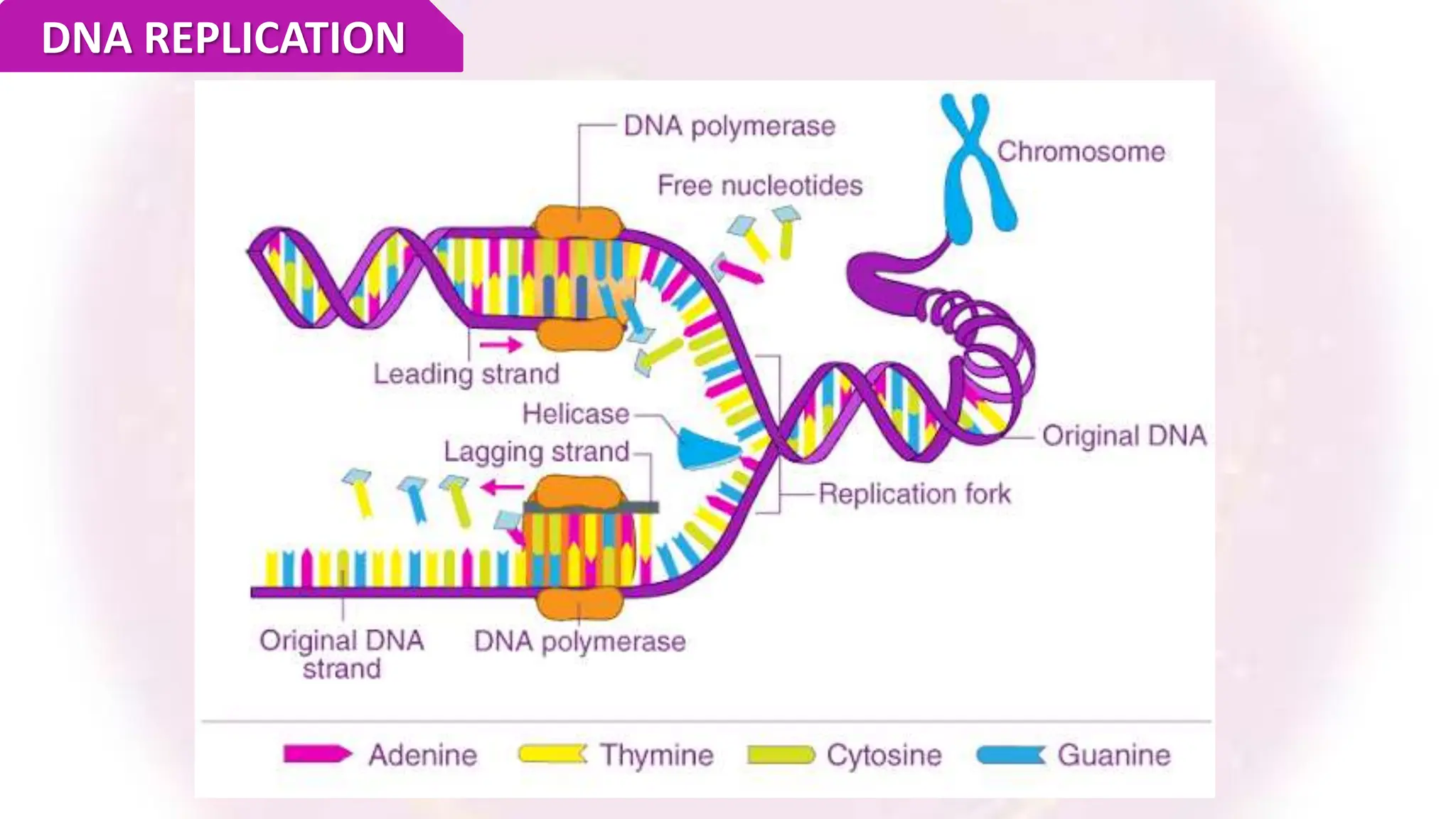
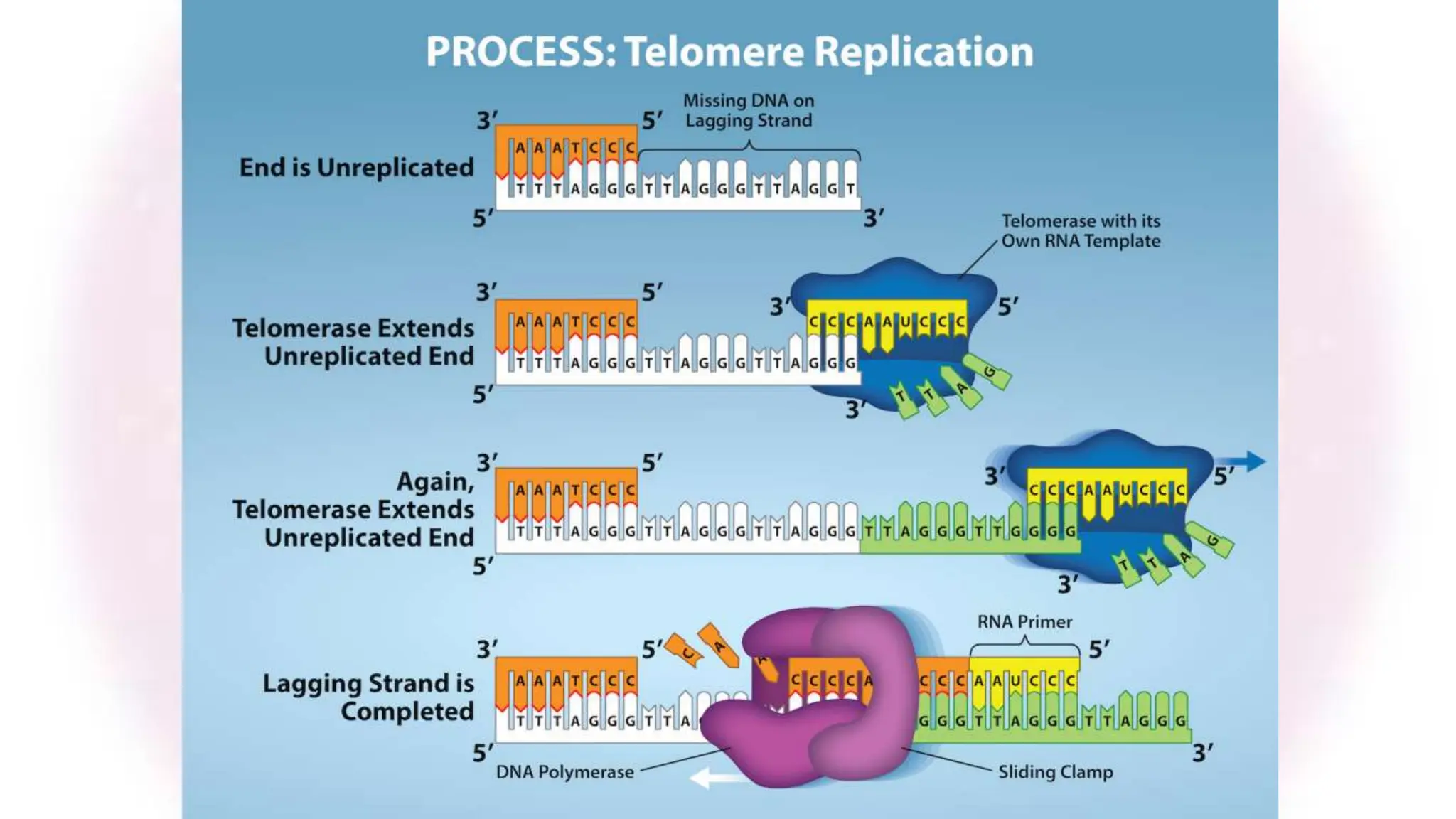
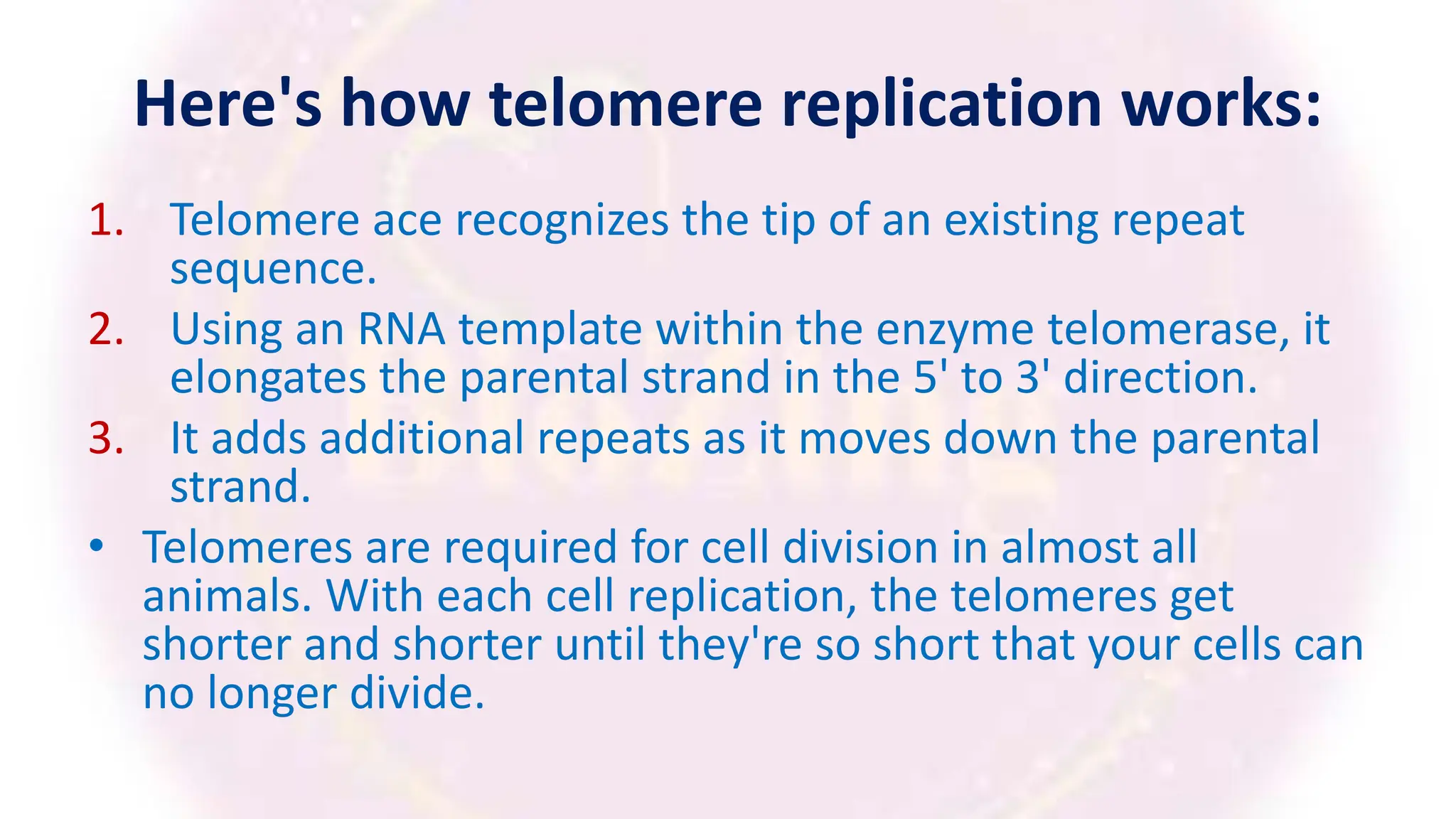
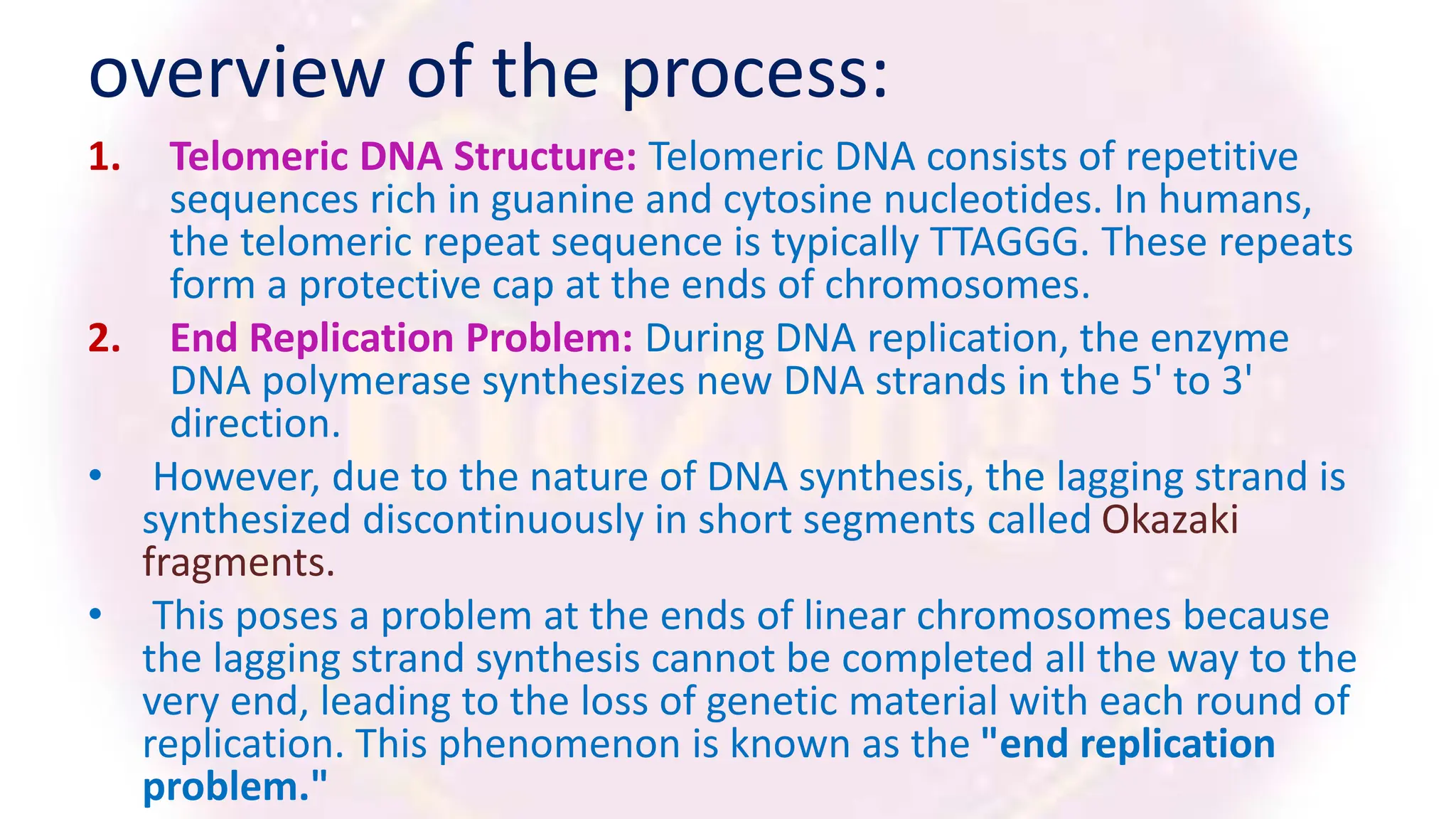
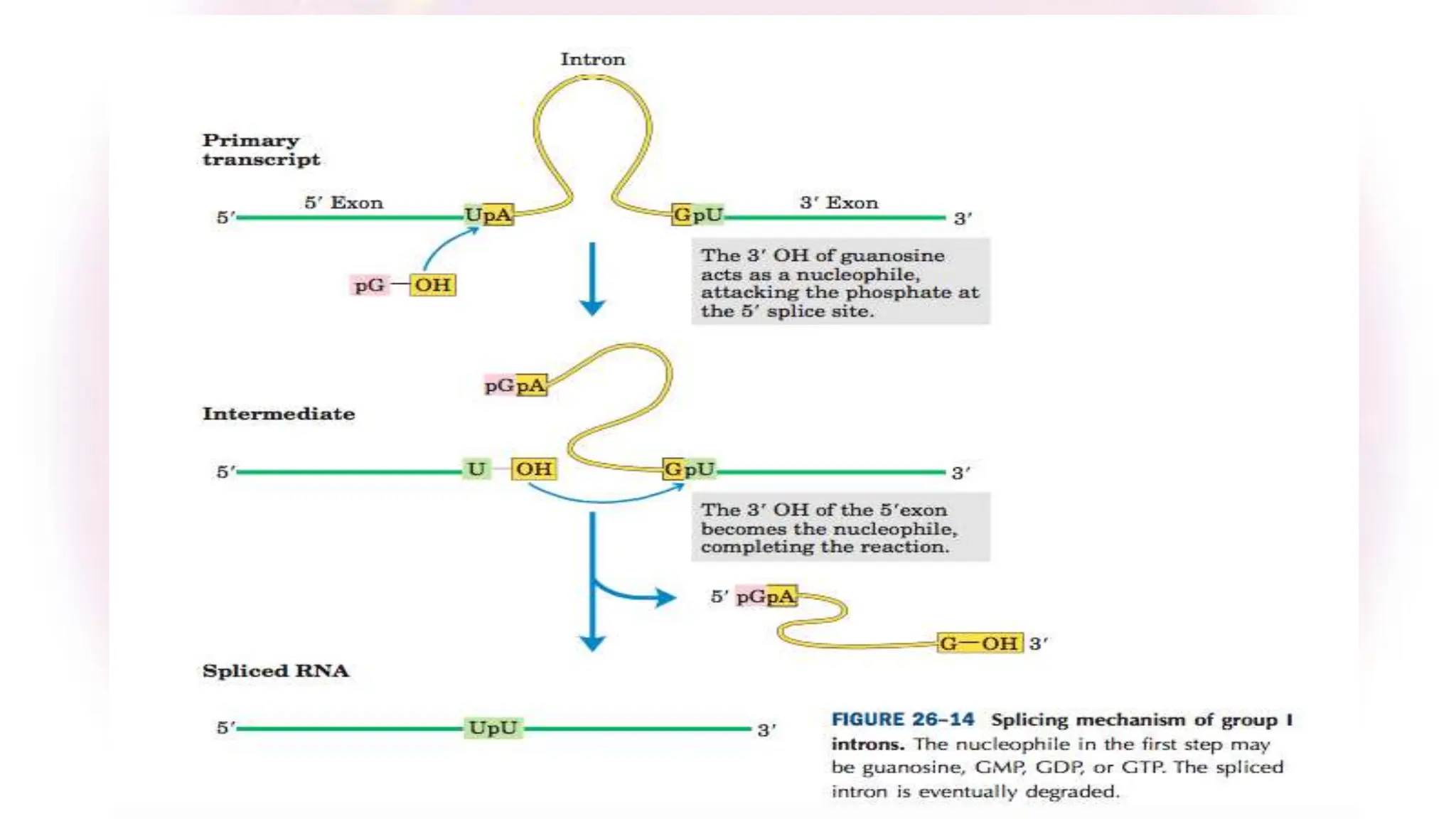
The document discusses key concepts in molecular biology, focusing on DNA replication in prokaryotes and eukaryotes, telomere replication, the features of the genetic code, and mRNA processing in eukaryotes. It details the mechanisms of DNA replication, such as initiation, elongation, and termination in prokaryotes, along with the function of telomerase in maintaining chromosome integrity in eukaryotes. Additionally, it outlines the splicing process of mRNA and highlights the properties and exceptions of the genetic code.