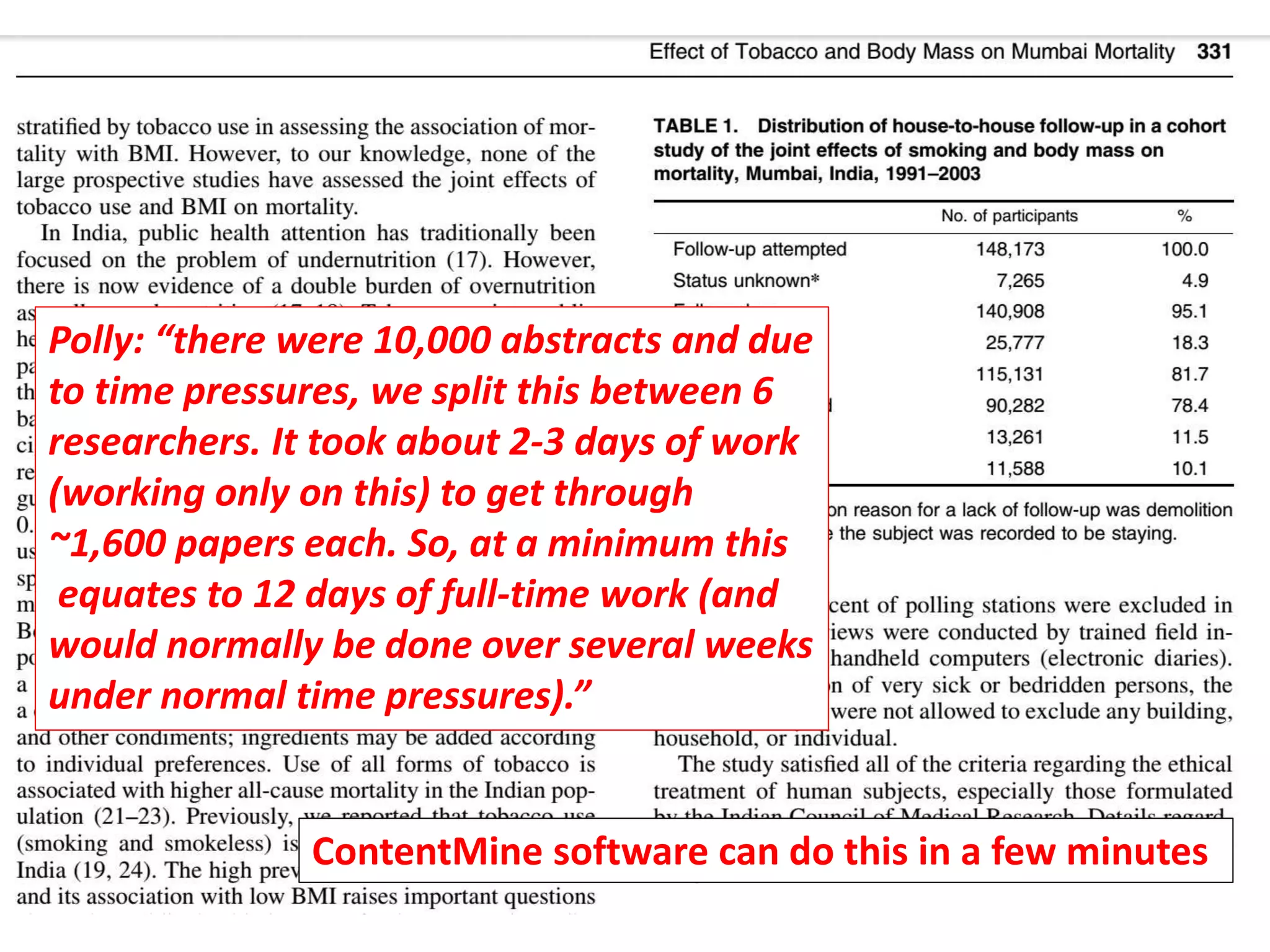
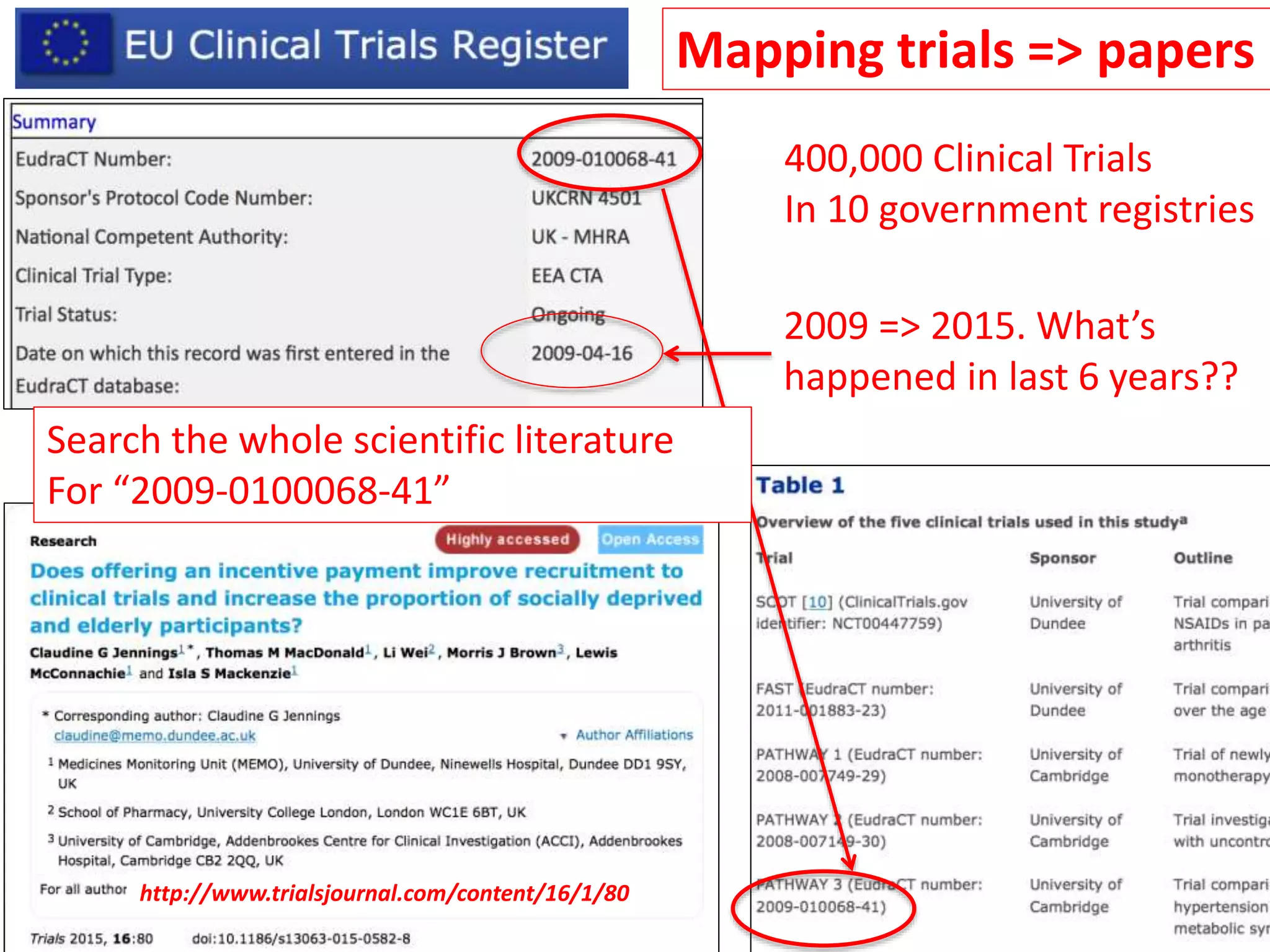
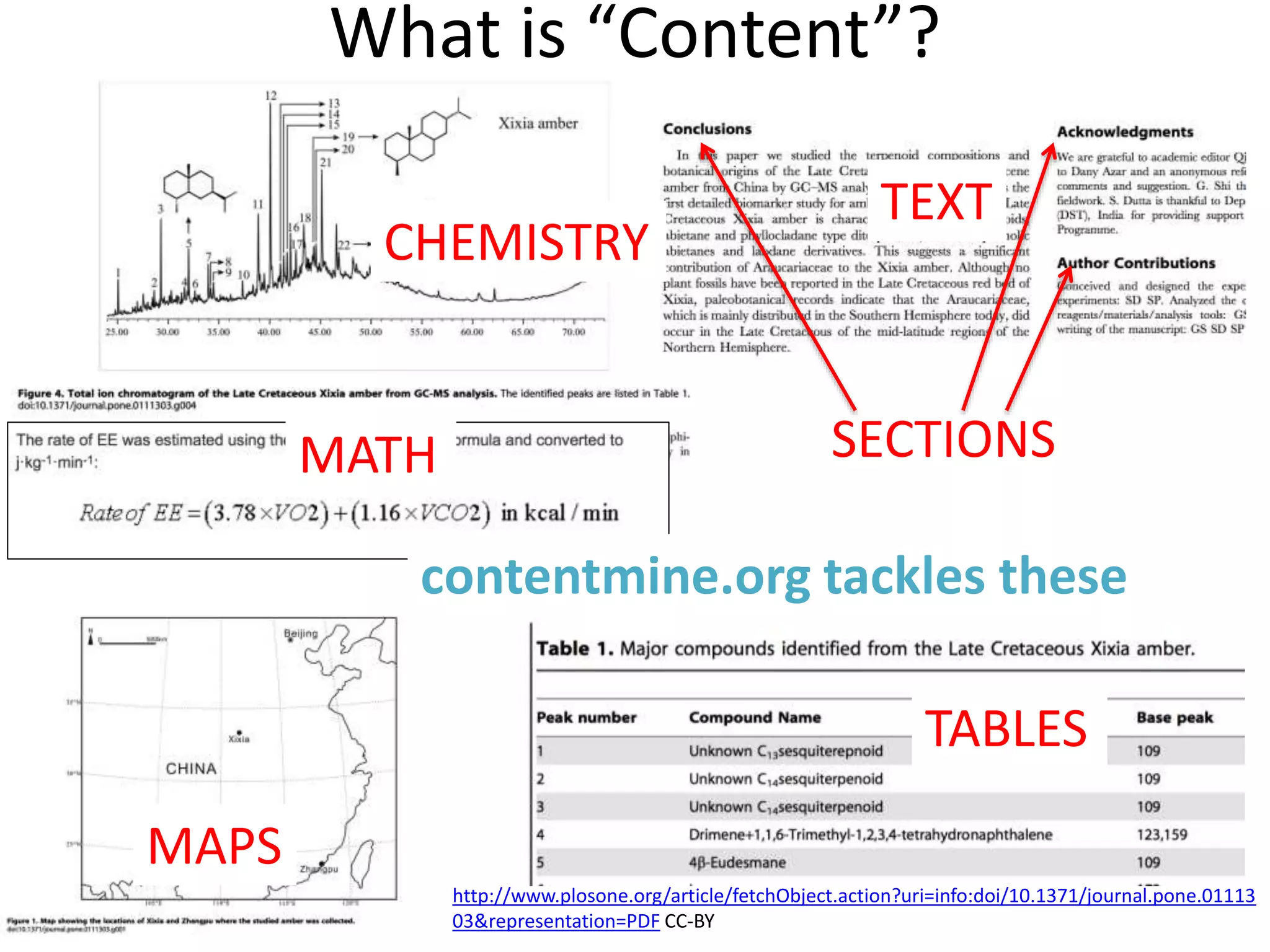
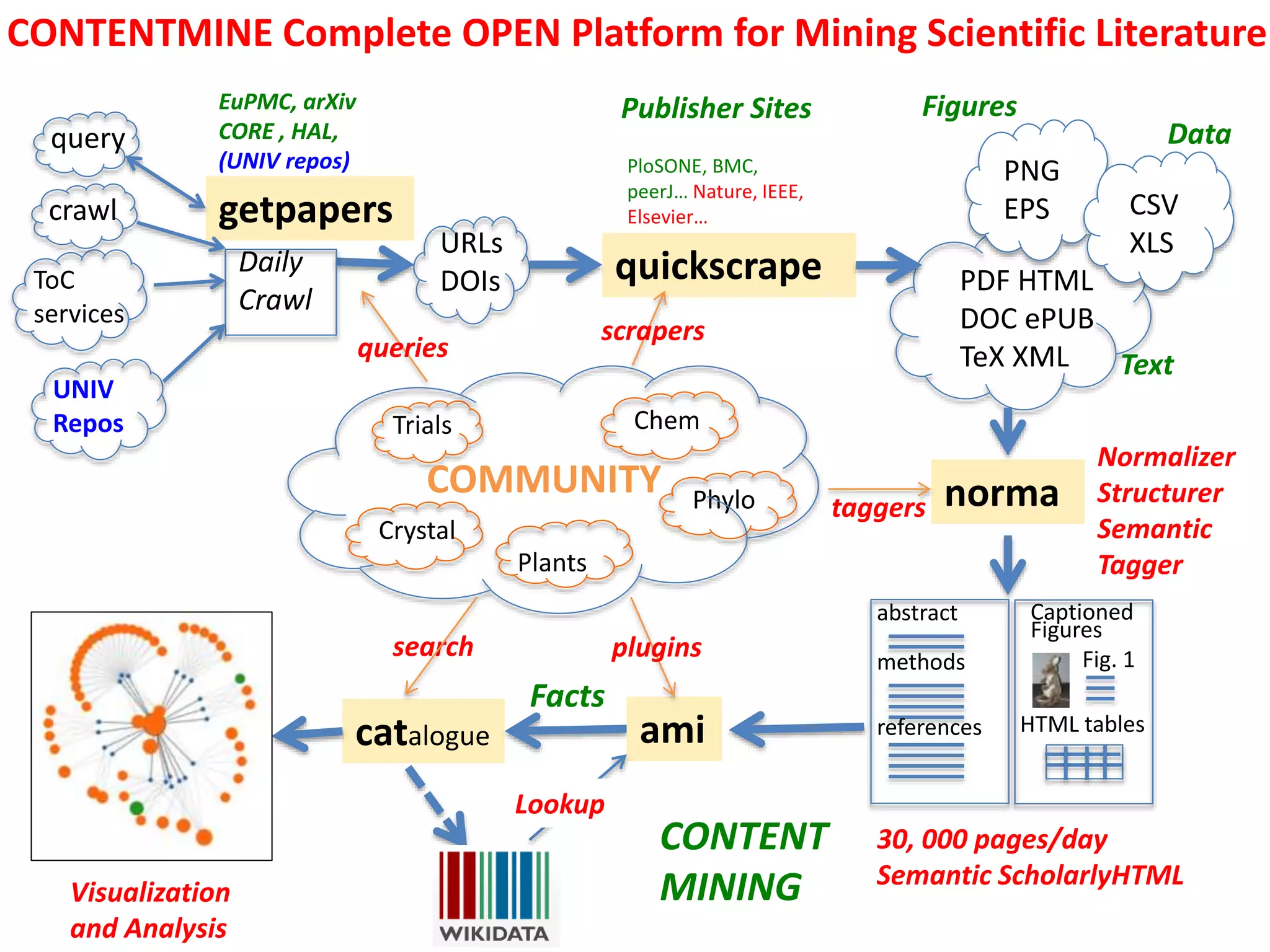
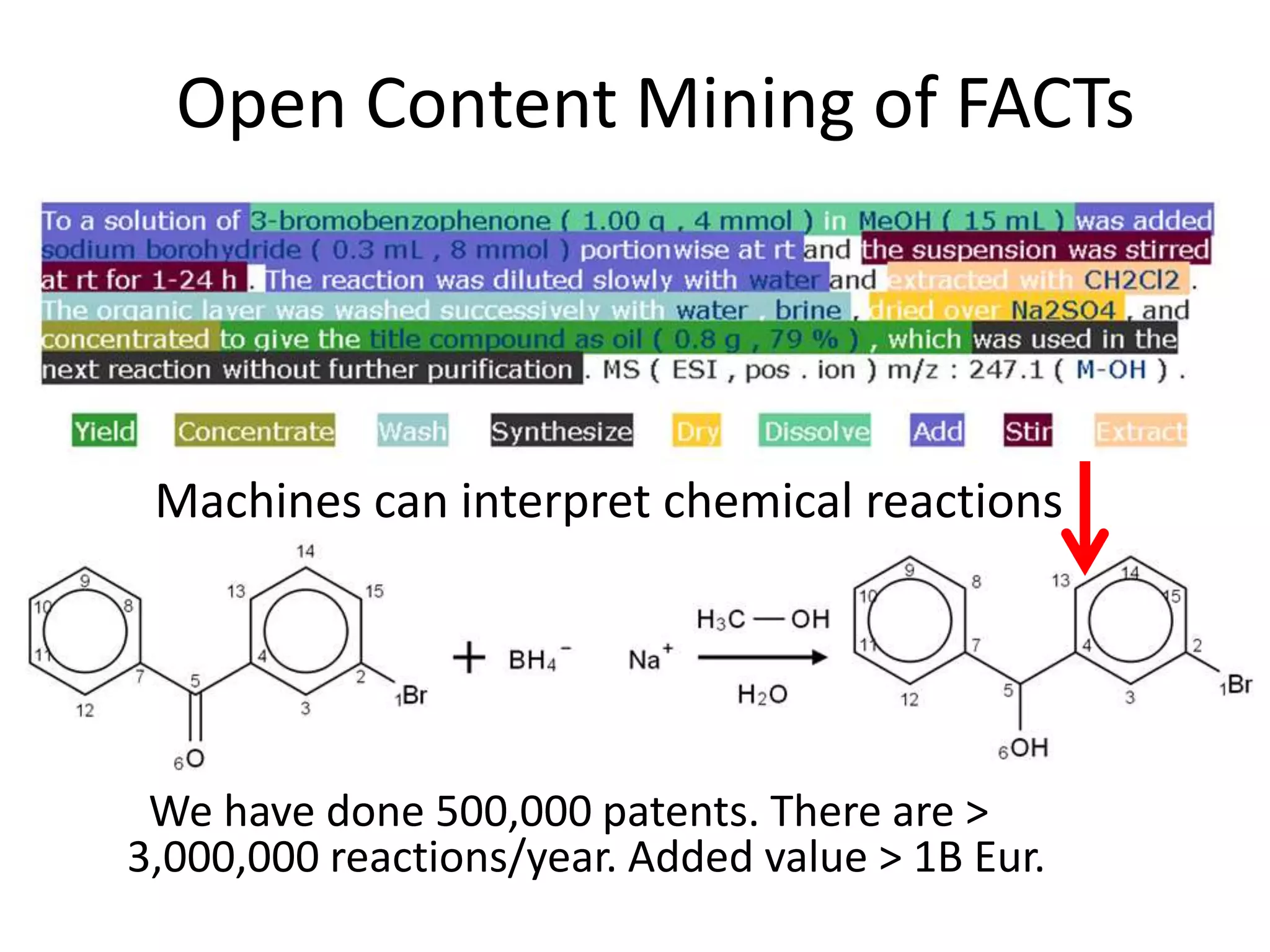
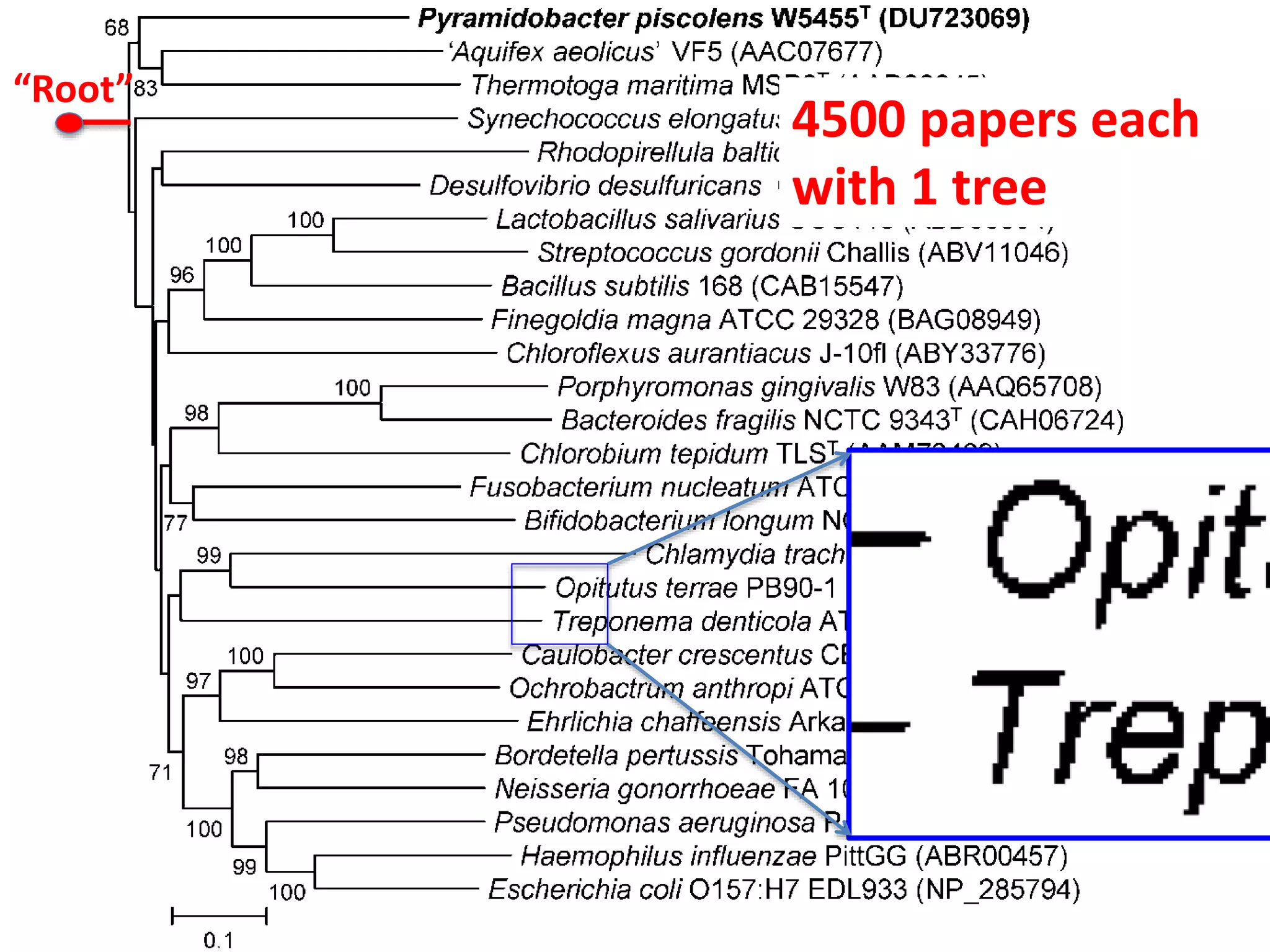
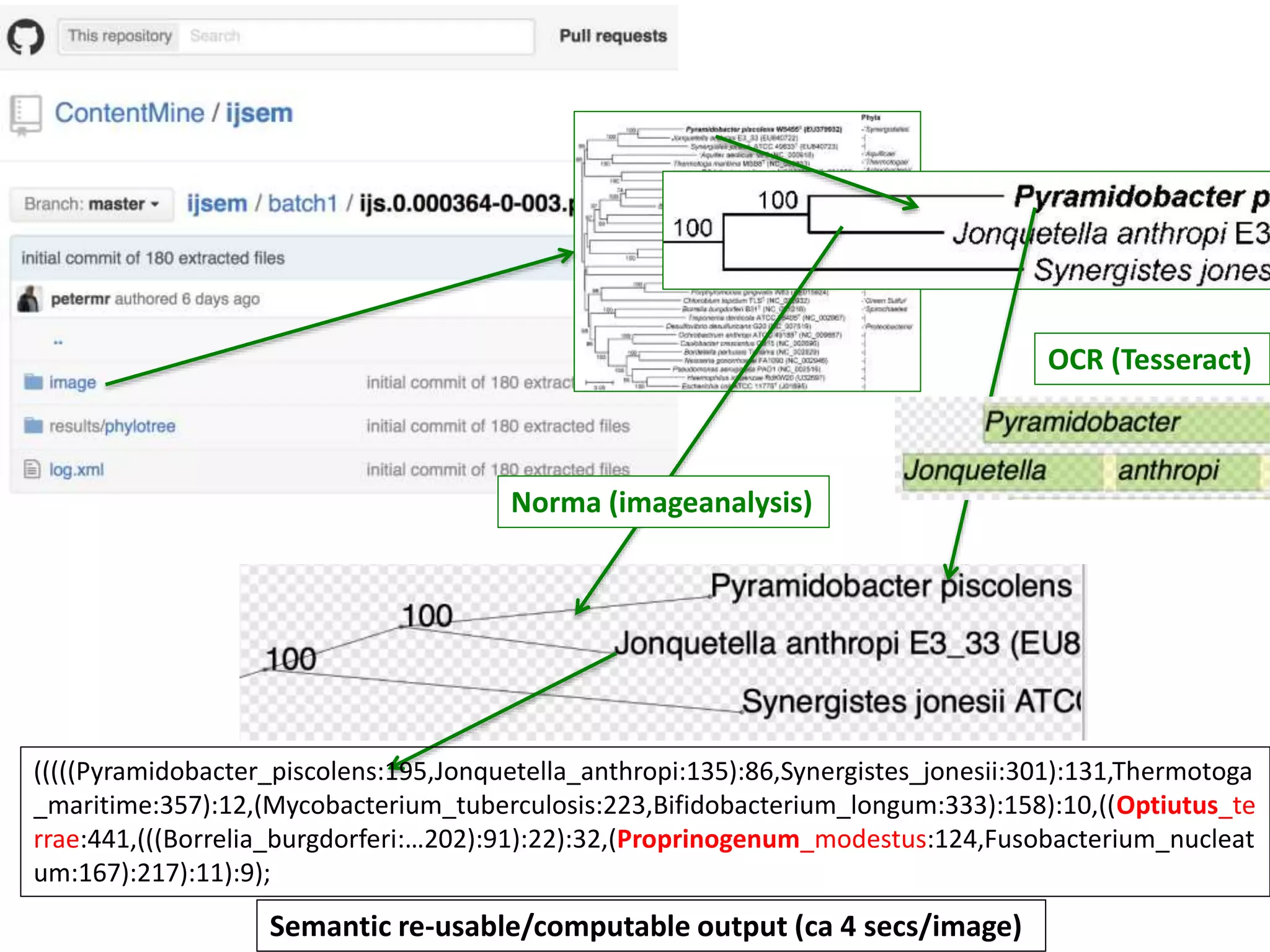
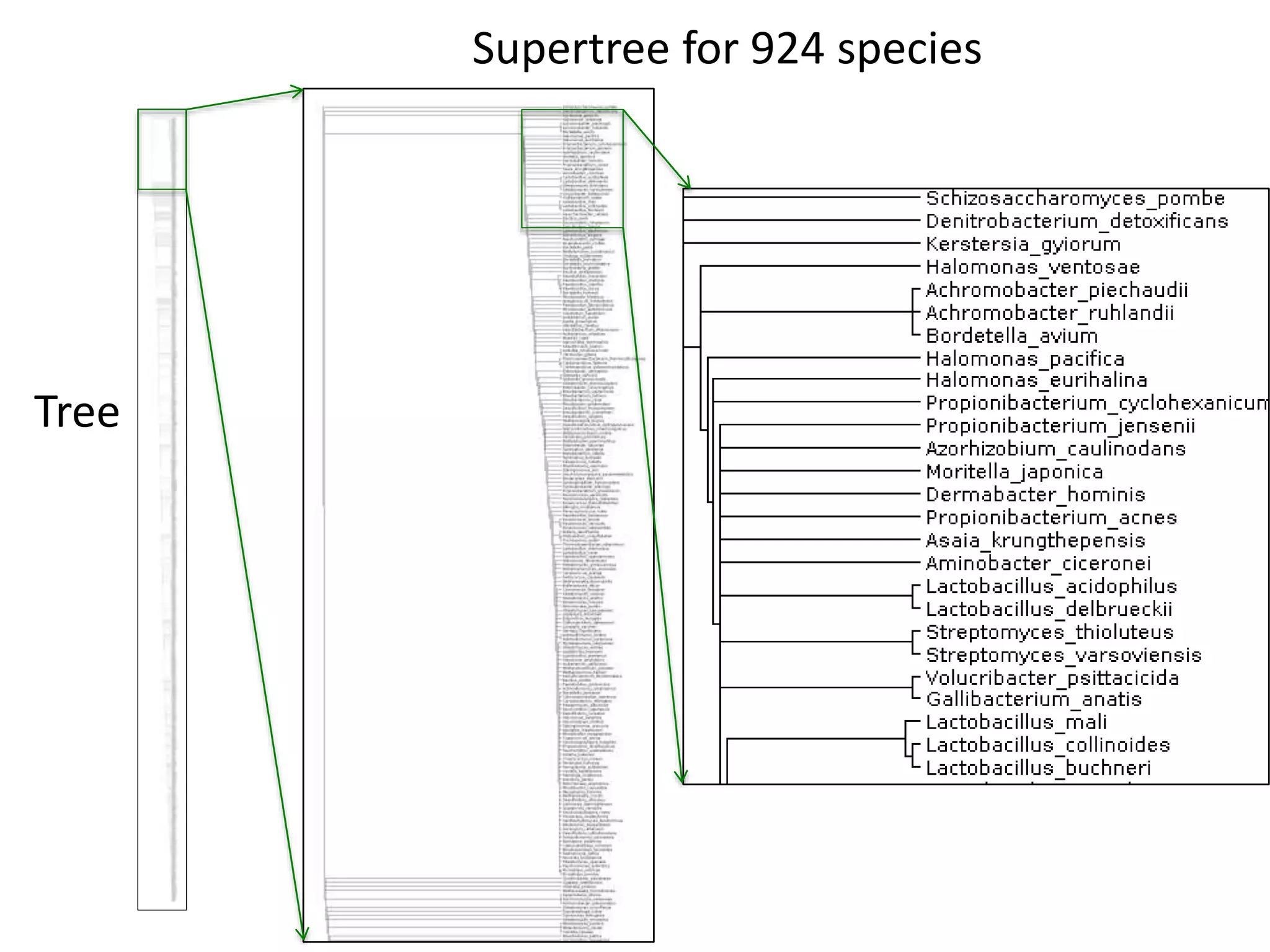
The document discusses content mining in Europe, emphasizing its importance for scientific research and the challenges posed by copyright and restrictive practices from publishers. It highlights various use cases, such as mapping clinical trials and extracting chemical reactions, showcasing how tools like ContentMine can significantly reduce the time needed for literature review. The document also addresses legal aspects regarding copyright and suggests that reproducible scientific mining often conflicts with publisher interests.









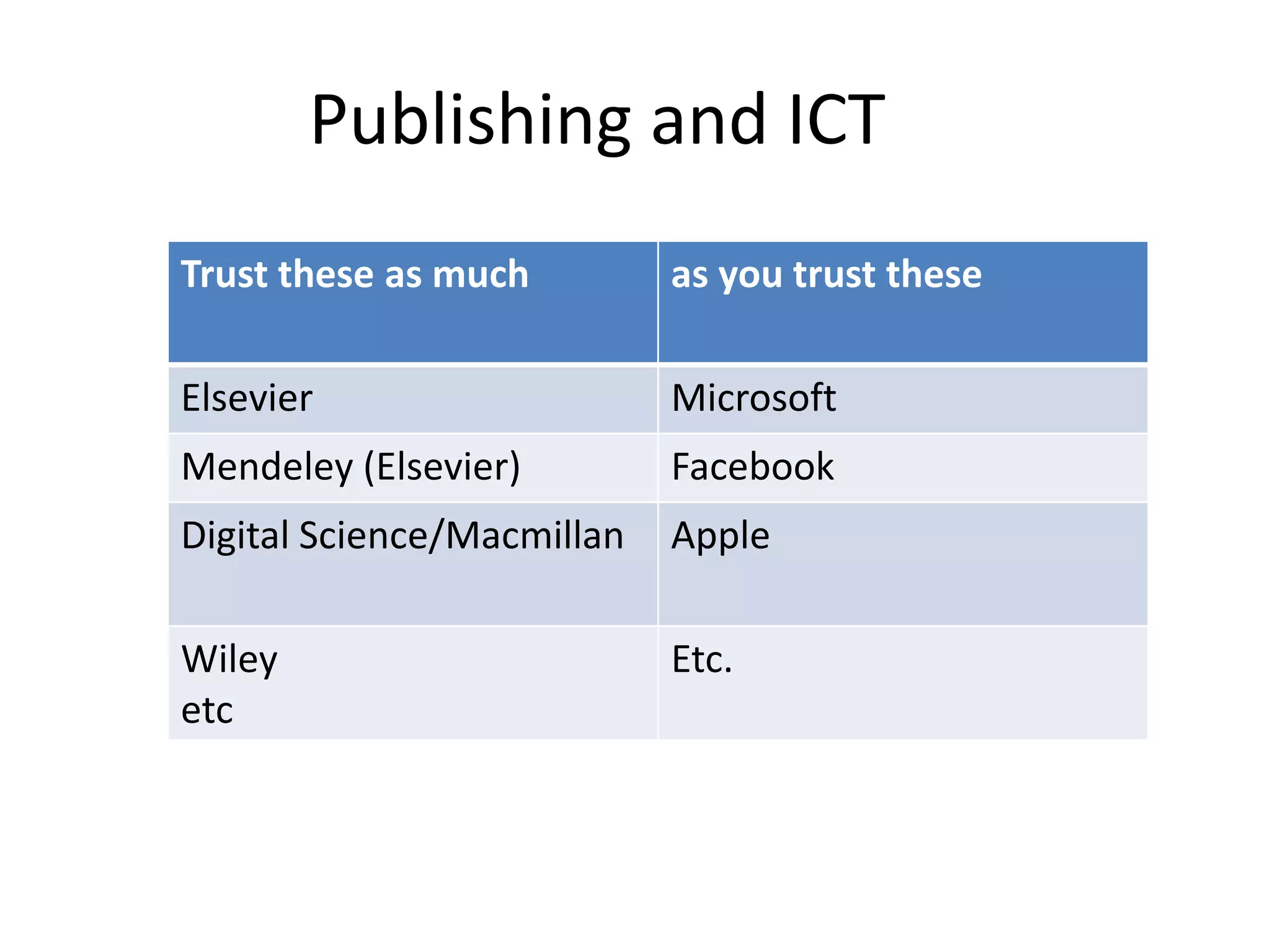
![STM Publishers prevent Mining
• FUD & disinformation about legality (Elsevier)
• Monopolies on infrastructure (“API”s, CCC
Rightfind)
• Technical obstruction (Wiley Captcha,
Macmillan Readcube)
• Restrictive contracts with libraries (ALL) [1]
• Wasting my/our time (ALL)
[1] [You may not] utilize the TDM Output to enhance … subject repositories
in a way that would [… ] have the potential to substitute and/or replicate
any other existing Elsevier products, services and/or solutions.](https://image.slidesharecdn.com/ofa2015-151022194657-lva1-app6891-151124140431-lva1-app6892/75/Content-Mining-of-Science-in-Europe-22-2048.jpg)
![WILEY … “new security feature… to prevent systematic download of content
“[limit of] 100 papers per day”
“essential security feature … to protect both parties (sic)”
CAPTCHA
User has to type words](https://image.slidesharecdn.com/ofa2015-151022194657-lva1-app6891-151124140431-lva1-app6892/75/Content-Mining-of-Science-in-Europe-23-2048.jpg)
