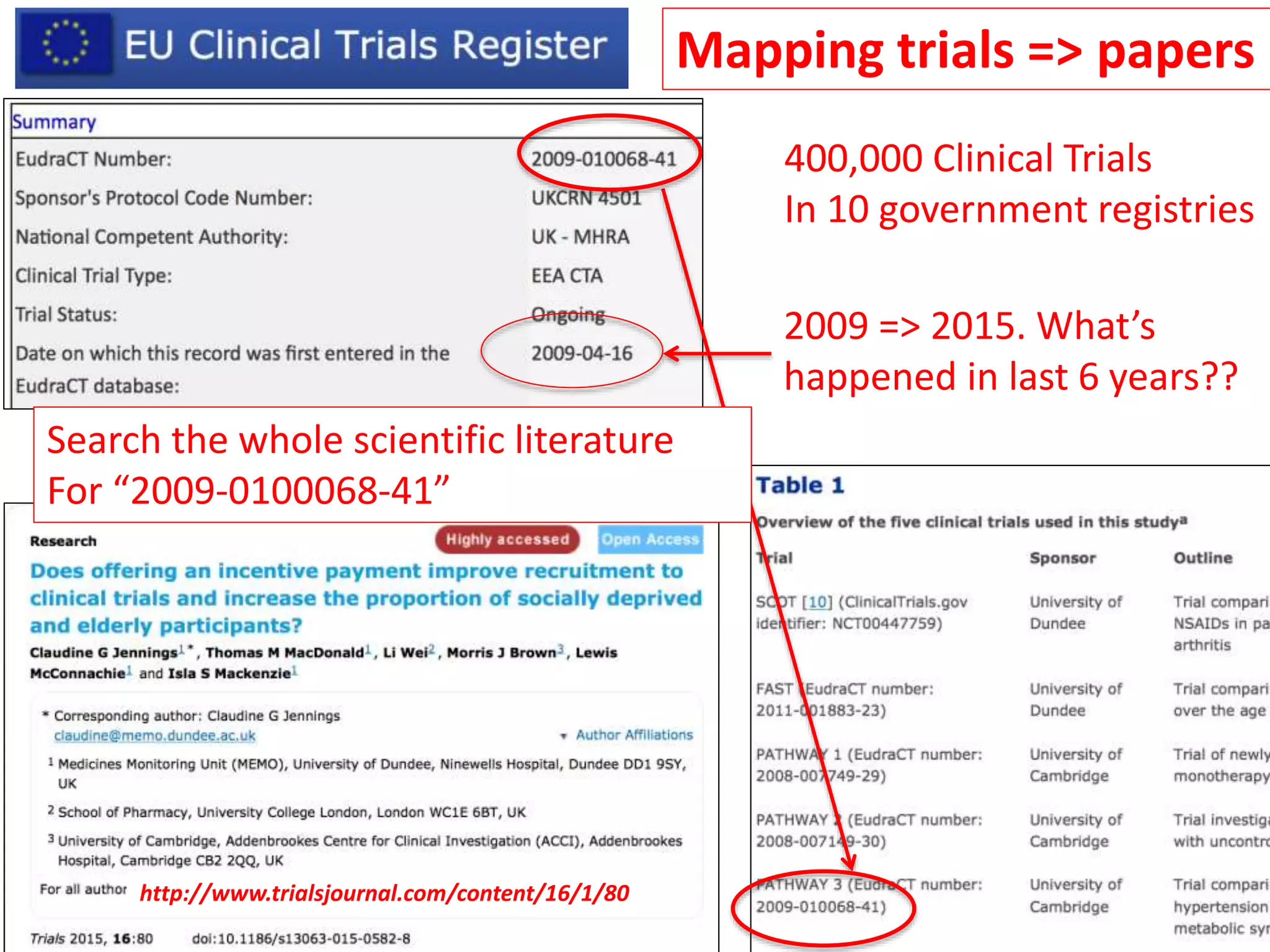
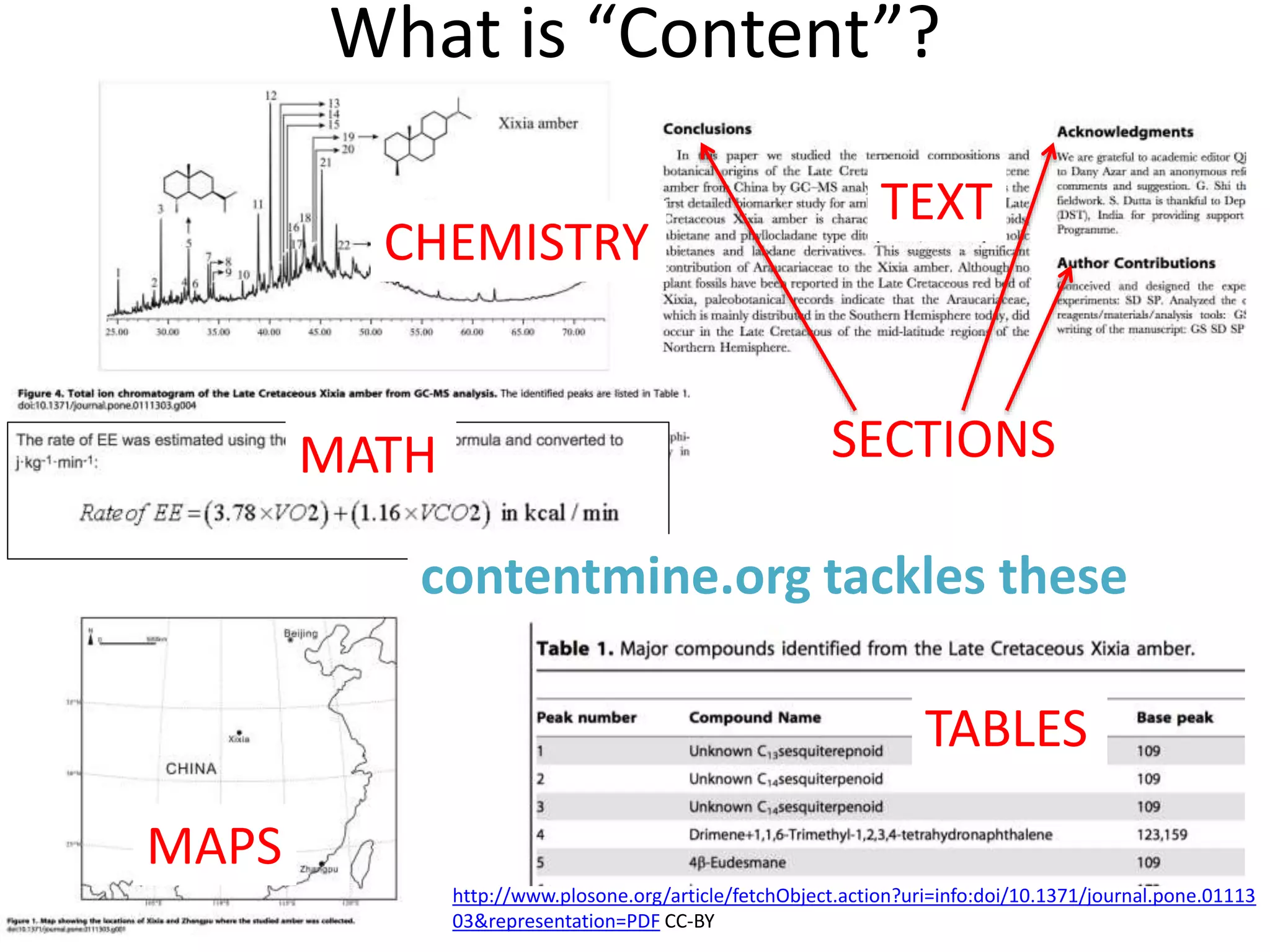
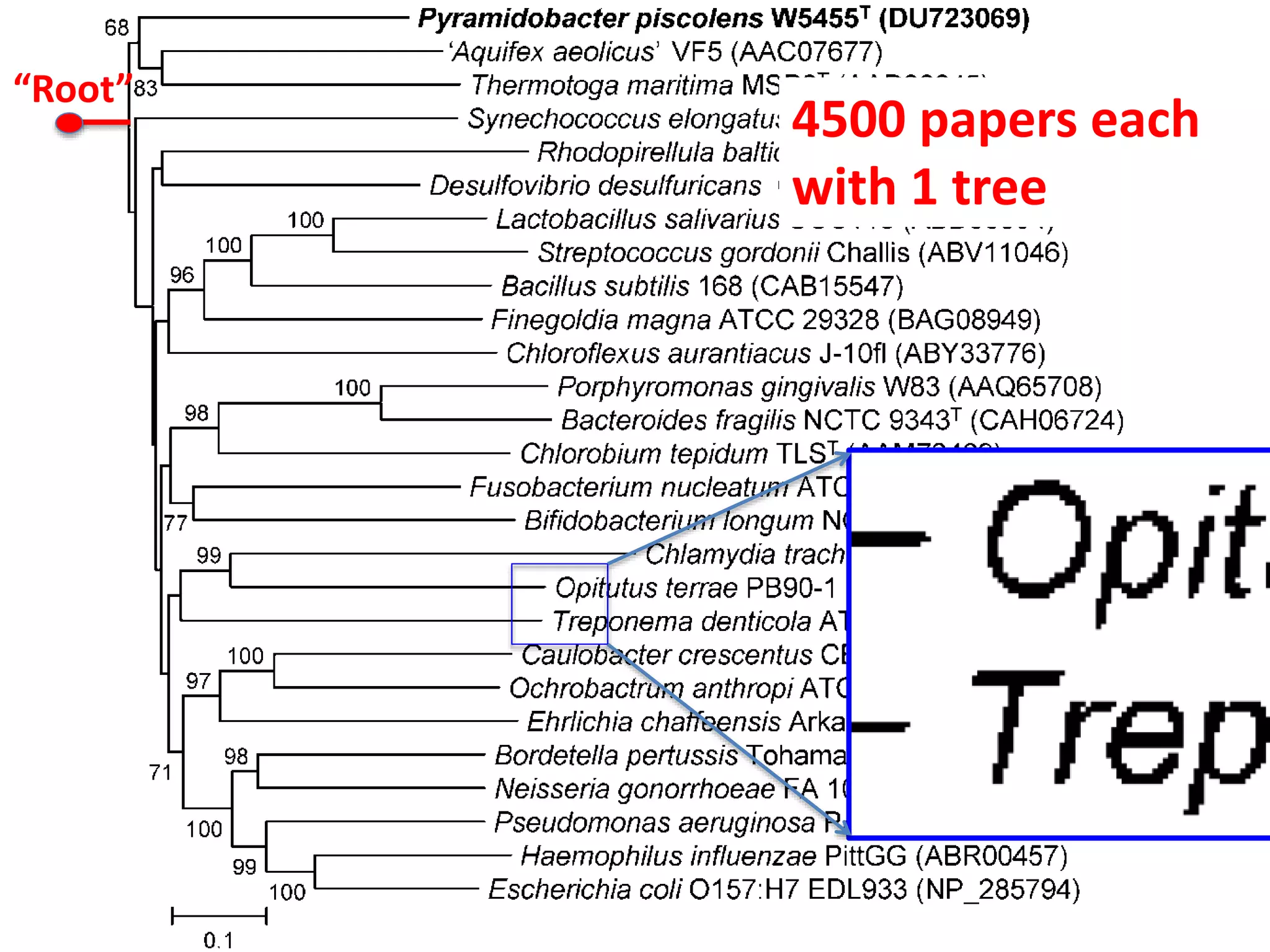
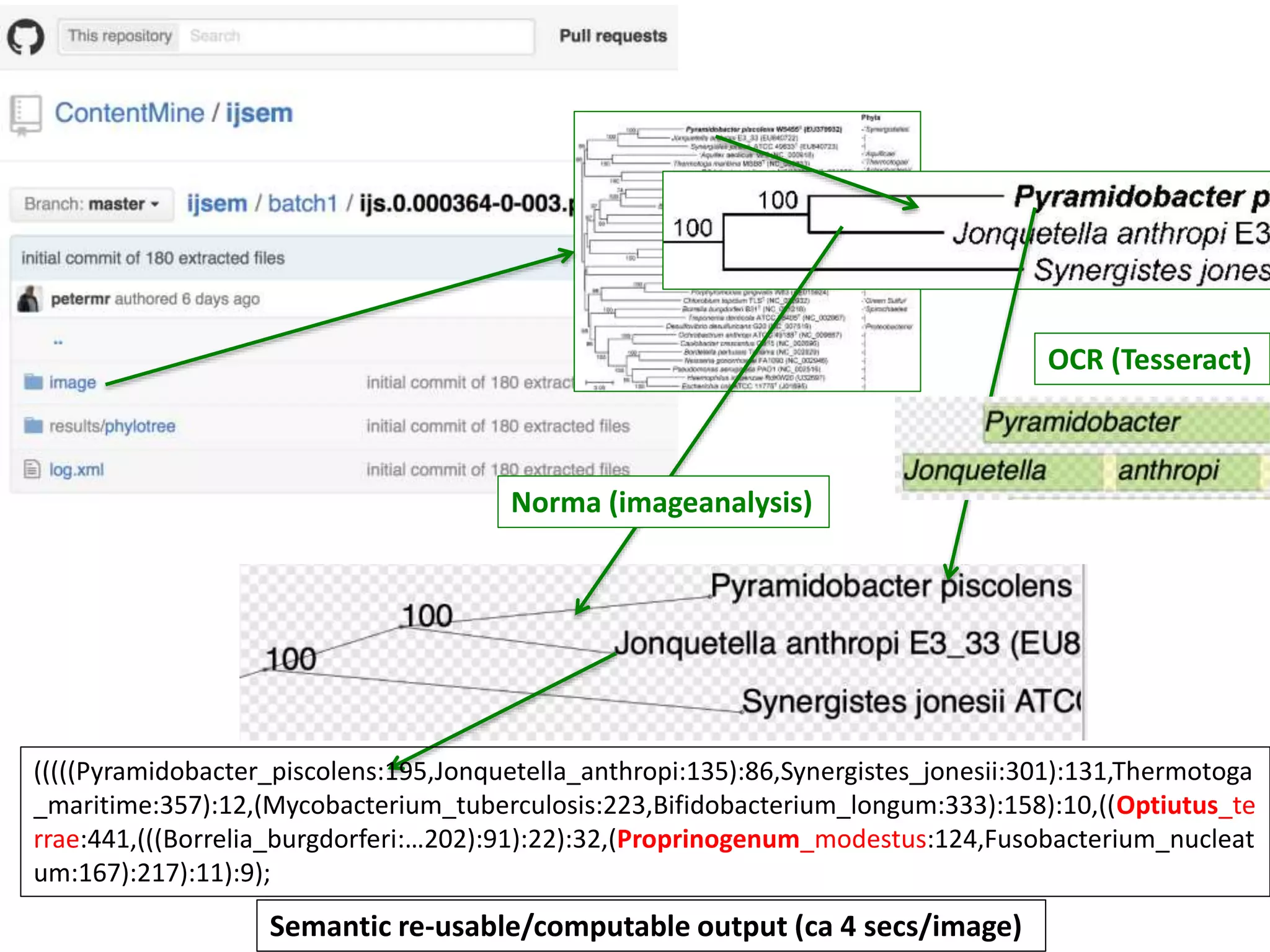
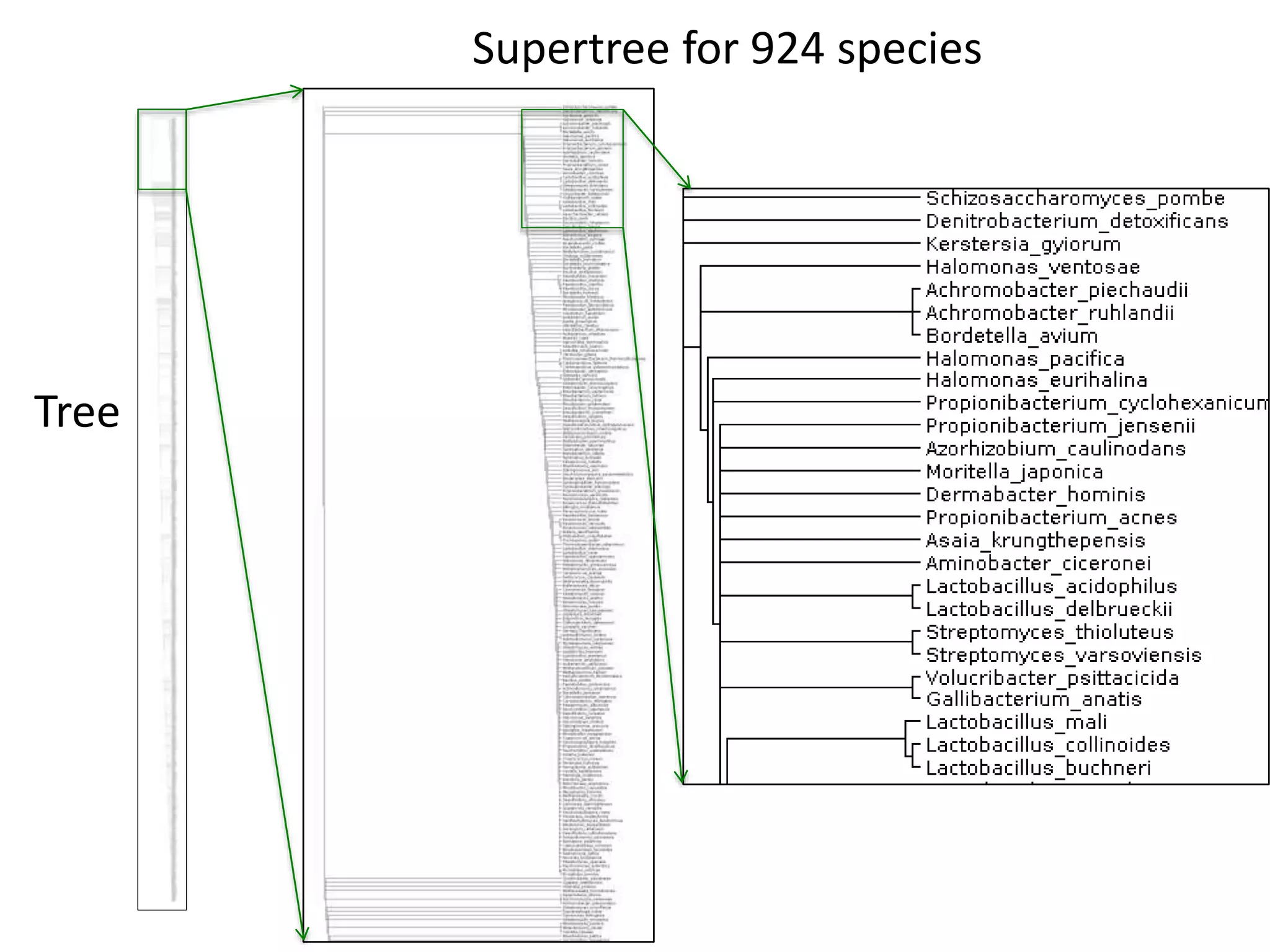
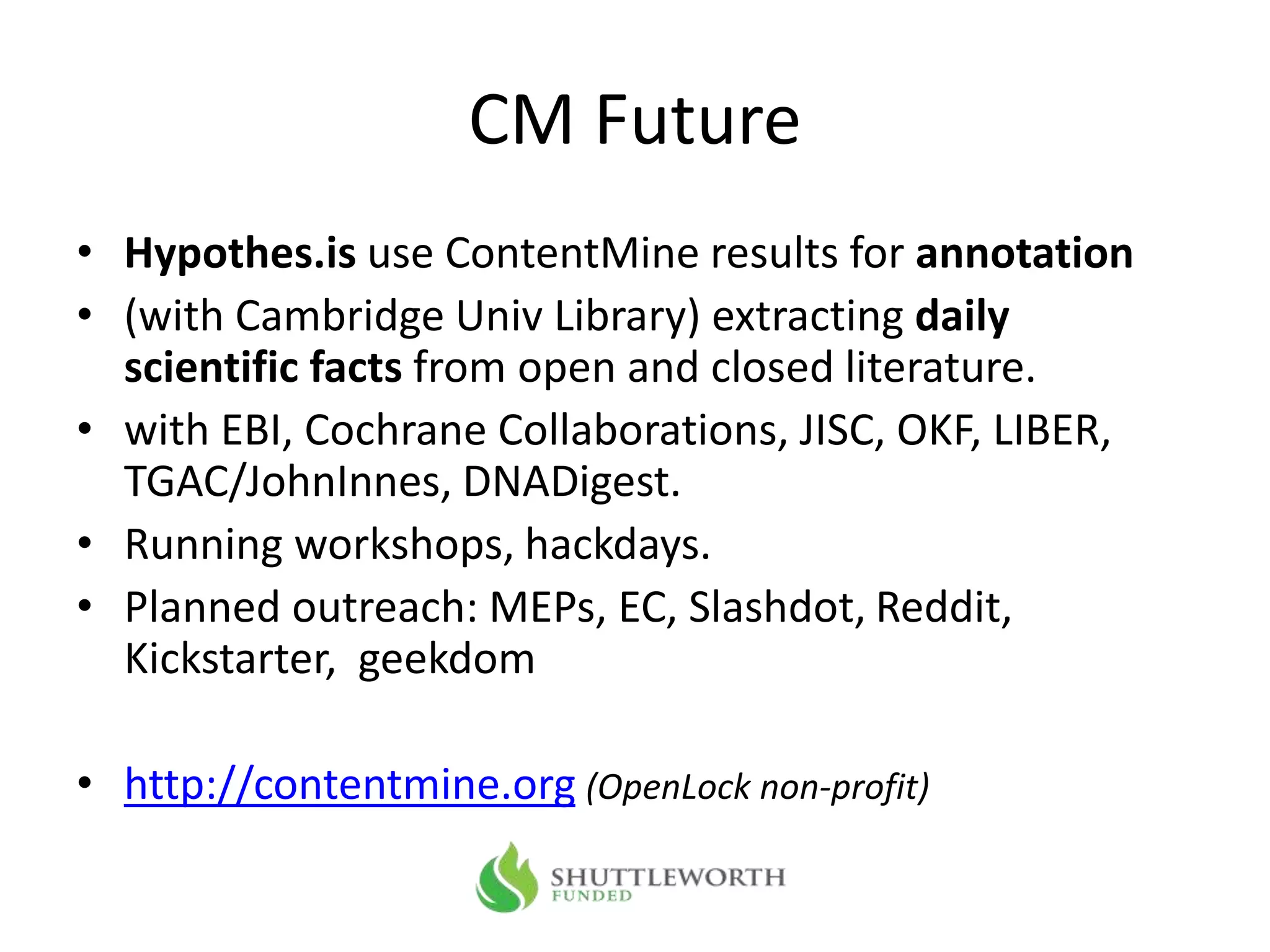
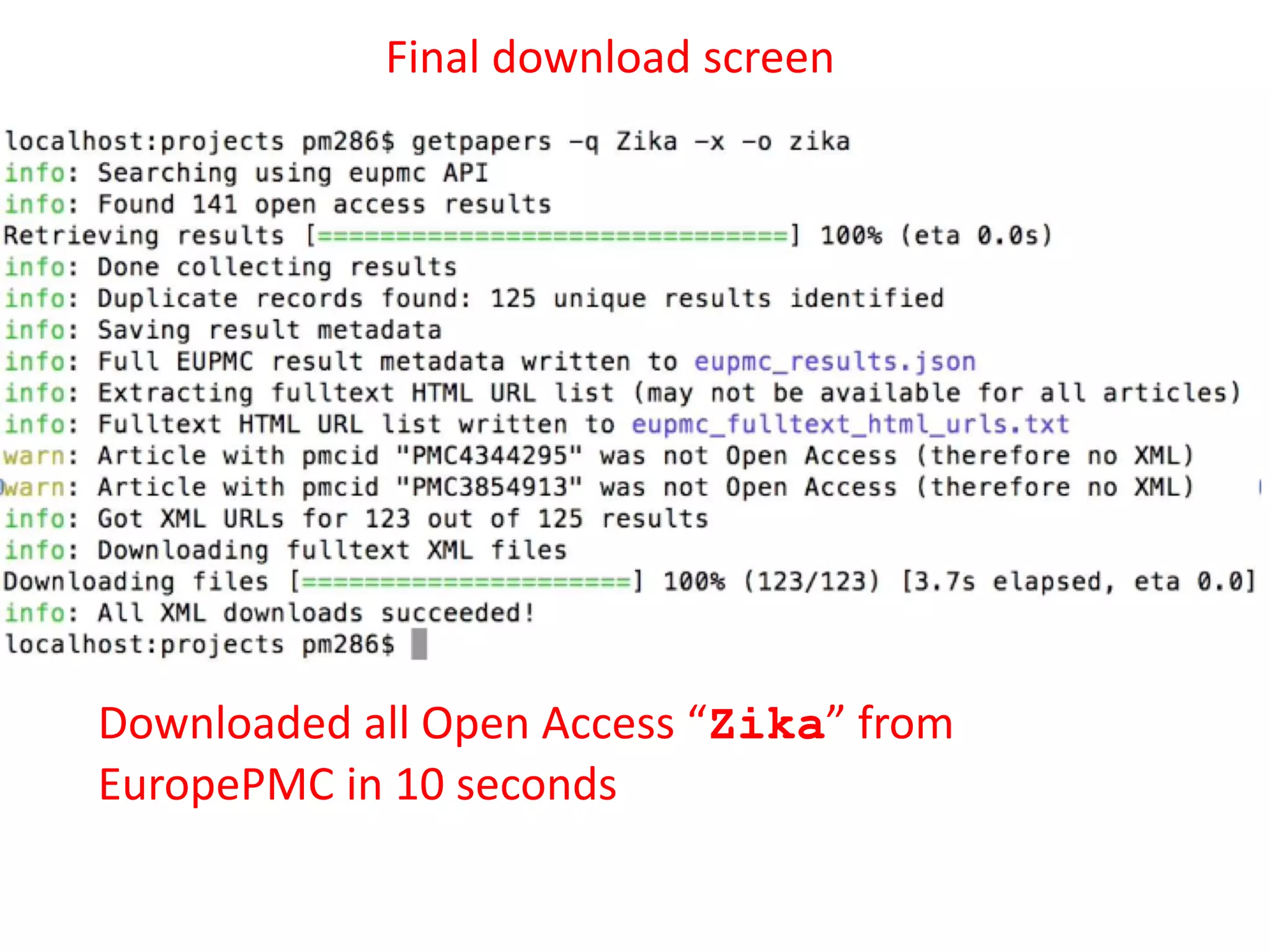
The document outlines the content mining strategies and tools developed by ContentMine to extract and analyze scientific literature, emphasizing the importance of open access and the right to mine data for research. It provides insight into the processing of scientific papers, enabling rapid access to essential information and trends in fields like biology and medicine. The presentation discusses various components of text and data mining, including crawling, scraping, and analyzing content from numerous sources.




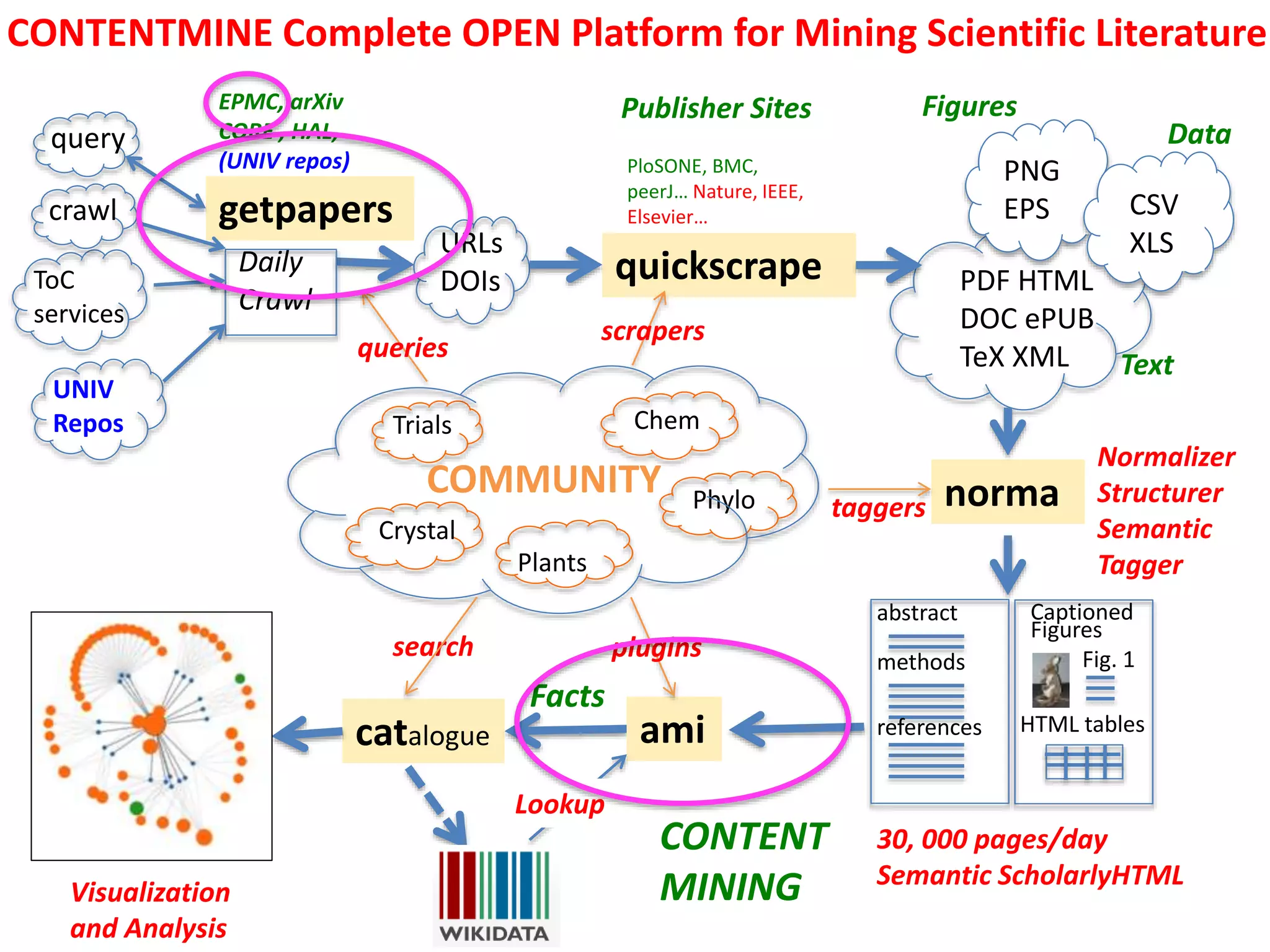
![Semantic Fulltext
• EuropePMC coherent OpenAccess
• getpapers: query , download (through API).
• AMI filters, checks[1], transforms facts in papers.
• sequences, species, genera, genes,
dictionaries
[0] All operations shown run in total of <3 minutes.
[1] Dictionaries and lookup.
[2] Usable from home by anyone
Zika endemic areas
Wikimedia CC-BY-SA](https://image.slidesharecdn.com/ftdmleiden-160229142301-160229153258/75/Content-Mining-of-Science-and-Medicine-6-2048.jpg)




![DNA Primers in running text
…the sodium channel voltage dependent gene (Nav). Primers
used to amplify this fragment were AaNaA
5’-ACAATGTGGATCGCTTCCC-3’
and AaNaB 5’-TGGACAAAAGCAAGGCTAAG-3’(8).
The primers amplify a fragment of approximately 472…
Snippet (quotable under 2014 UK Statutory Instrument (“Hargreaves”):
~/PMC4654492/results/sequence/dnaprimer/results.xml”
W3C Annotation
[PREFIX]
[MATCH] (link to target)
[SUFFIX]
CMine structure
plugin
option
DNA double stranded fragment
Wikimedia CC-BY-SA](https://image.slidesharecdn.com/ftdmleiden-160229142301-160229153258/75/Content-Mining-of-Science-and-Medicine-12-2048.jpg)