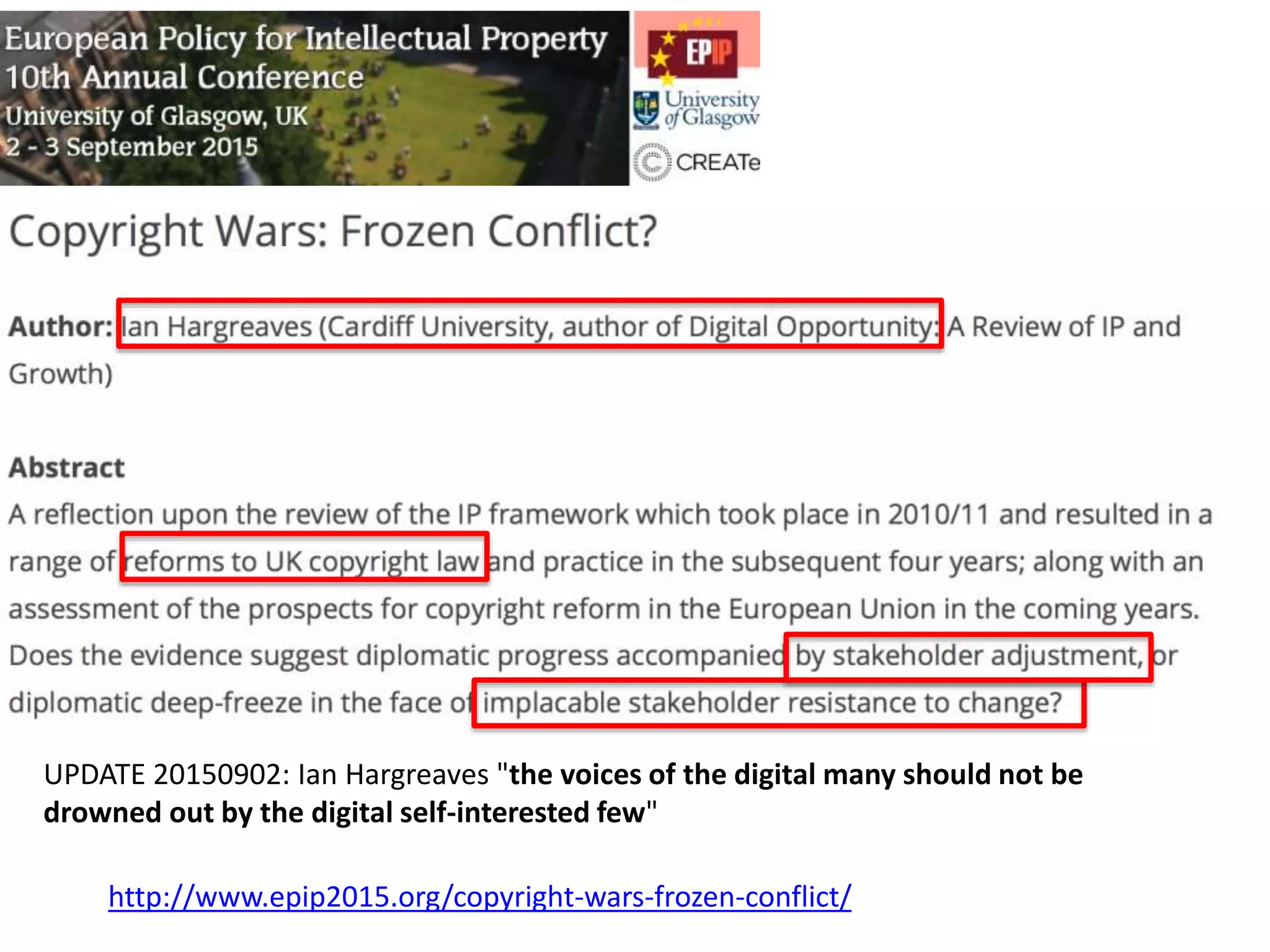
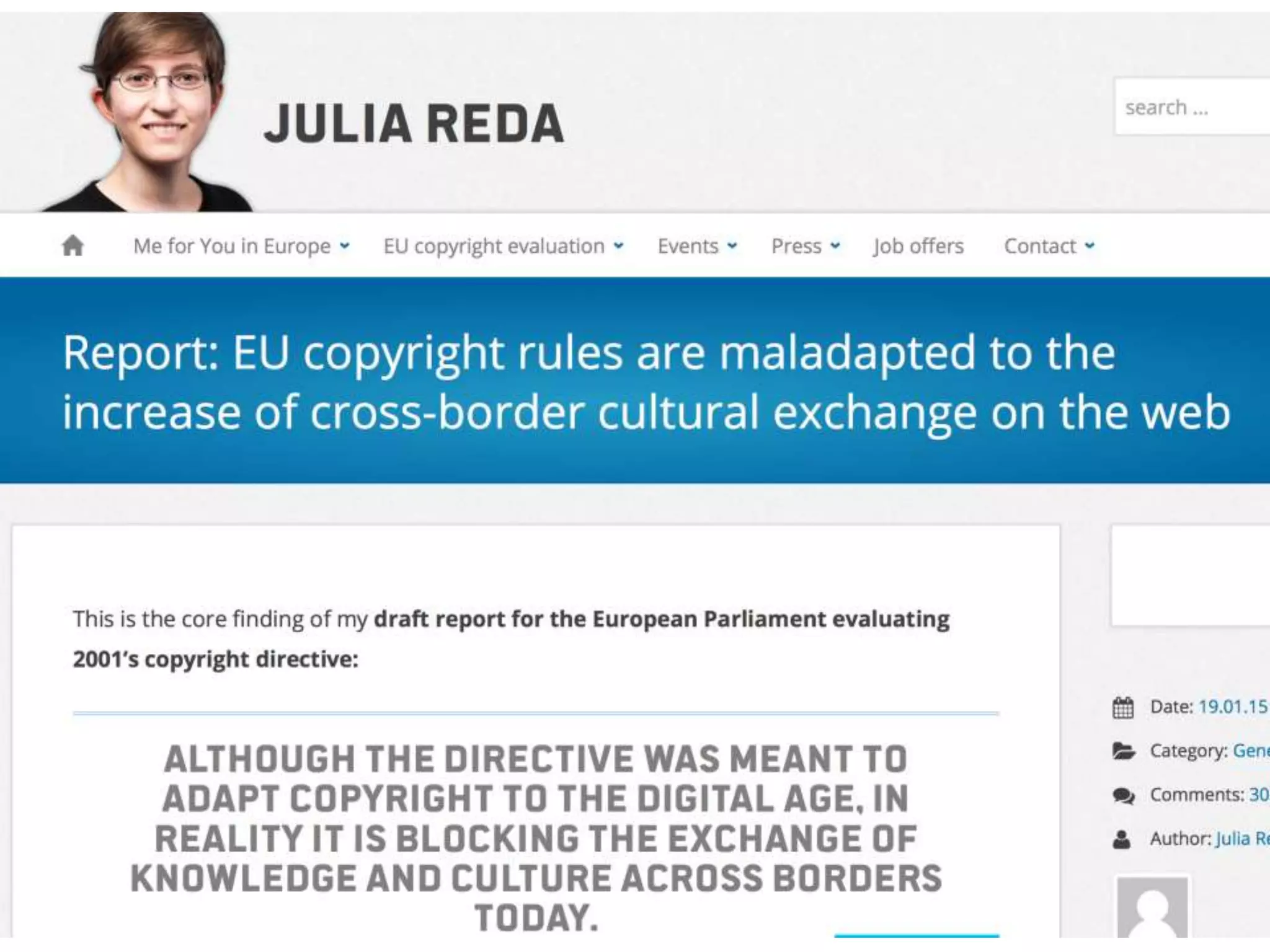
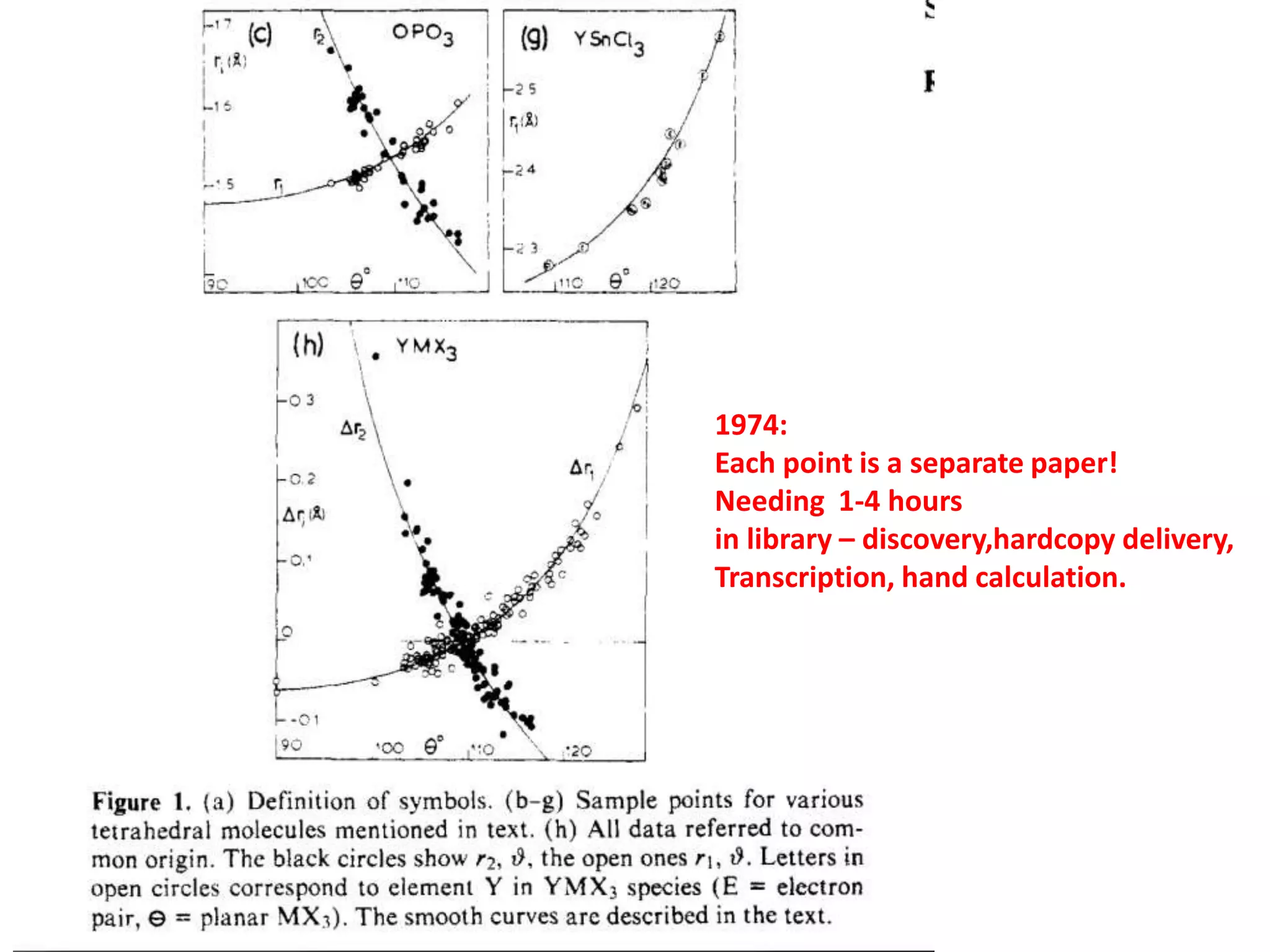
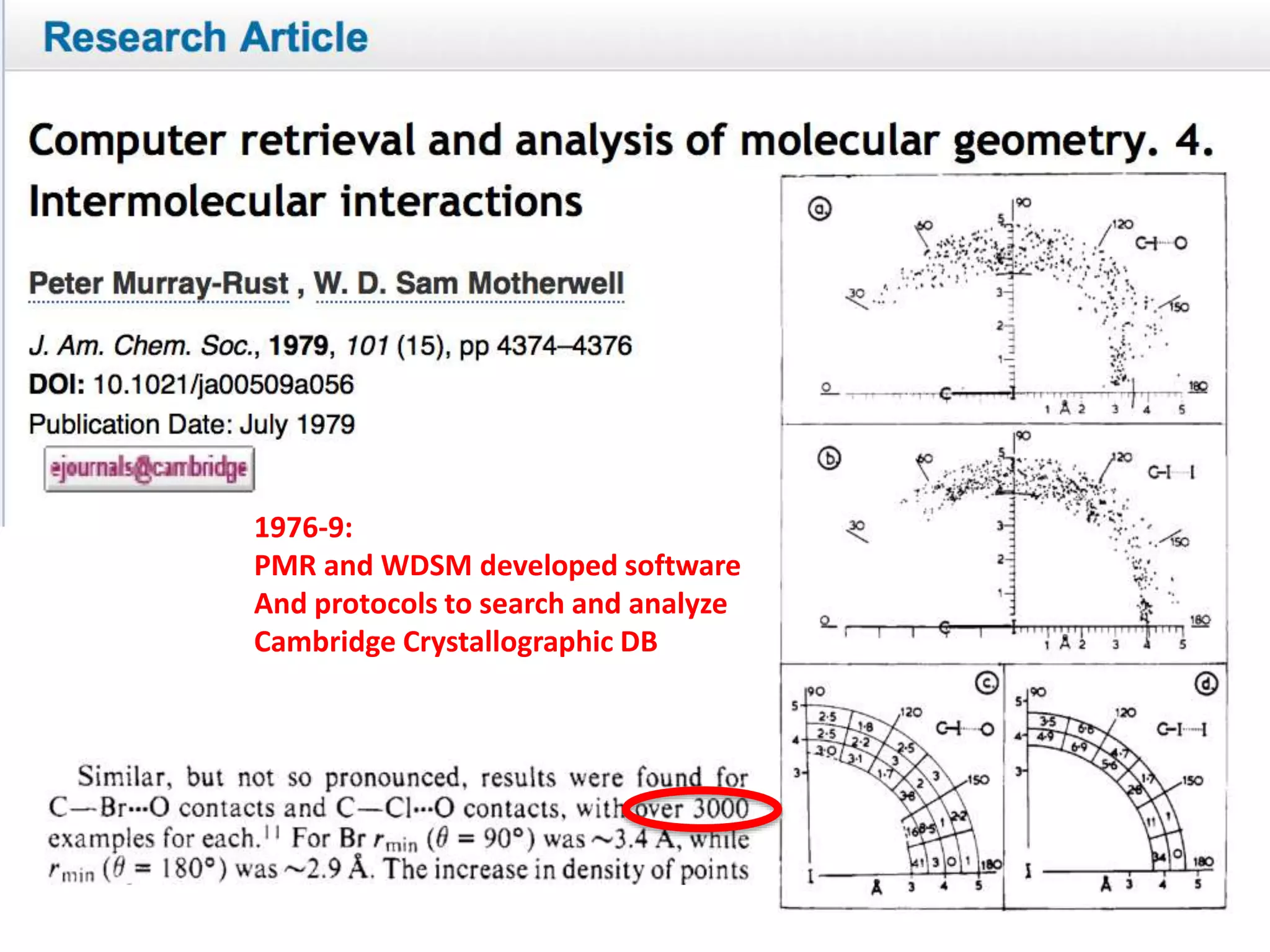
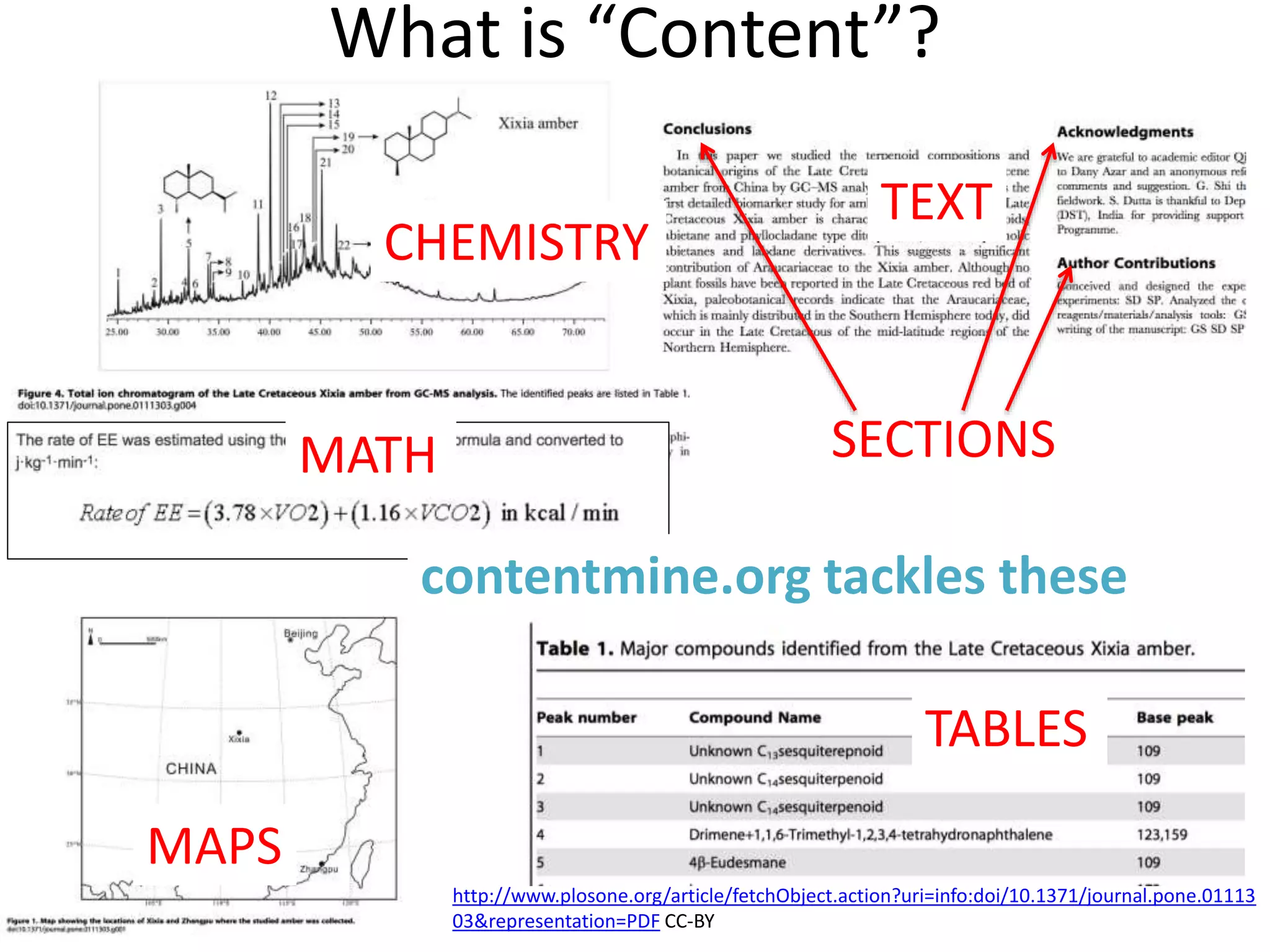
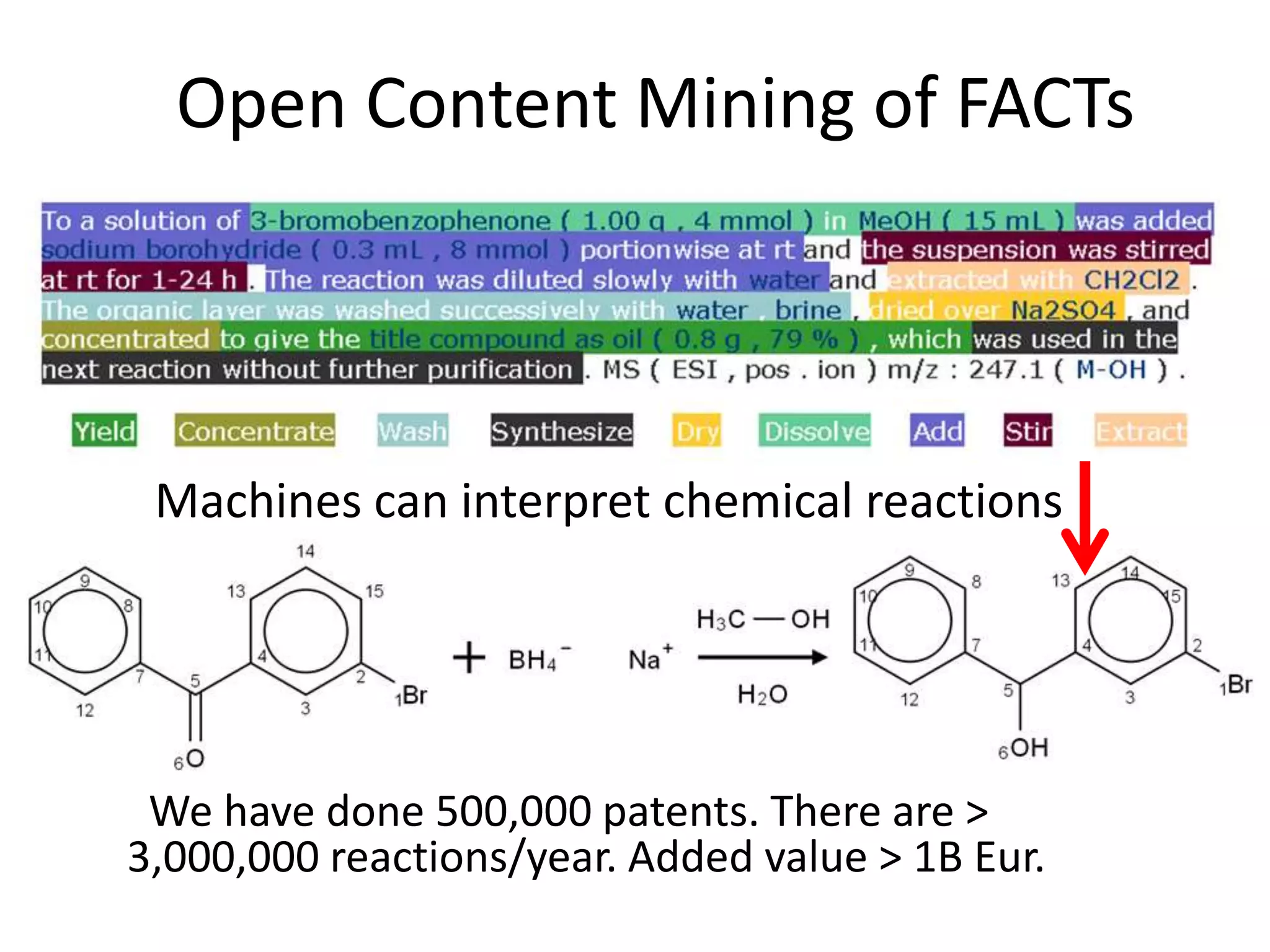
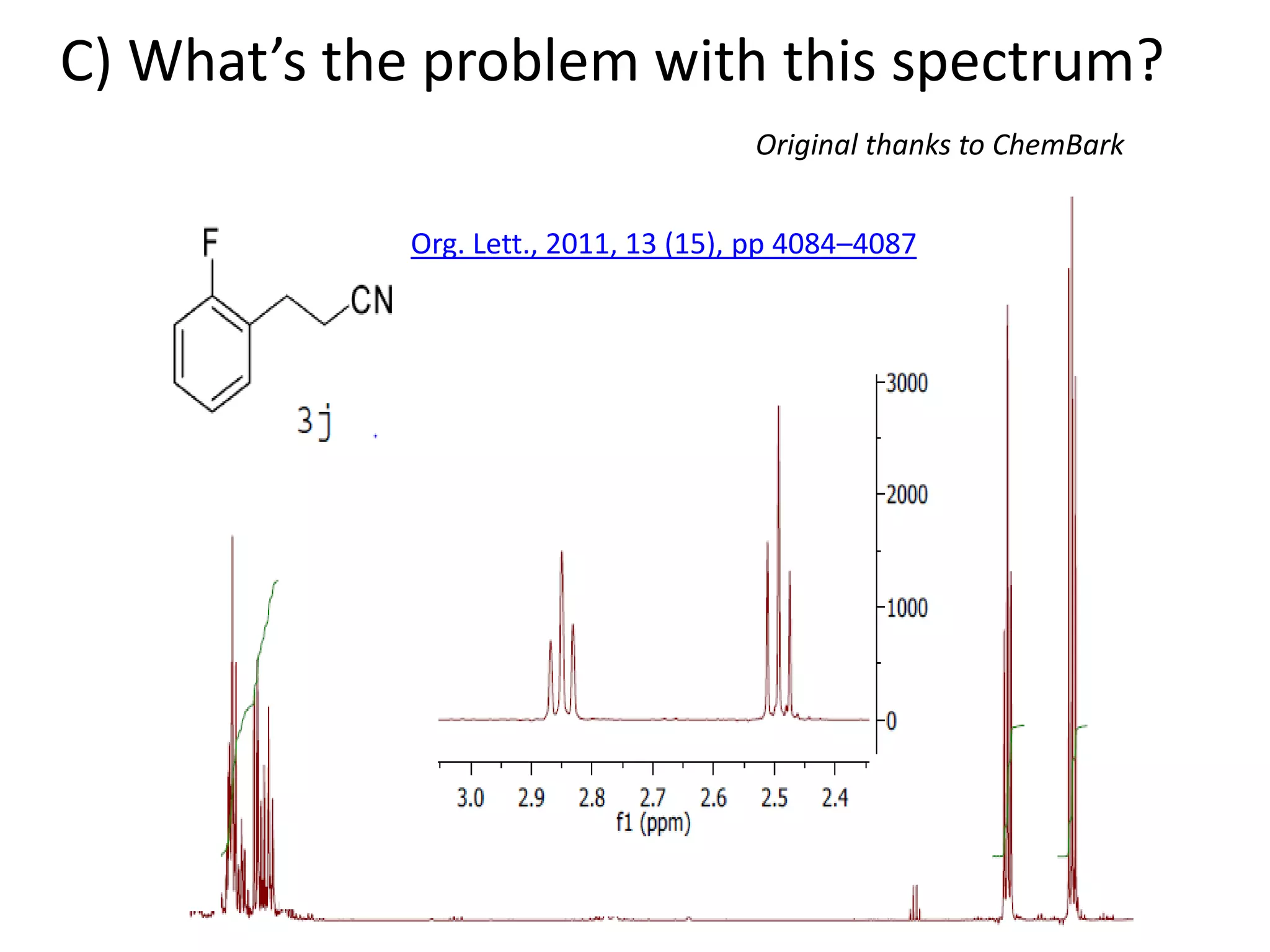
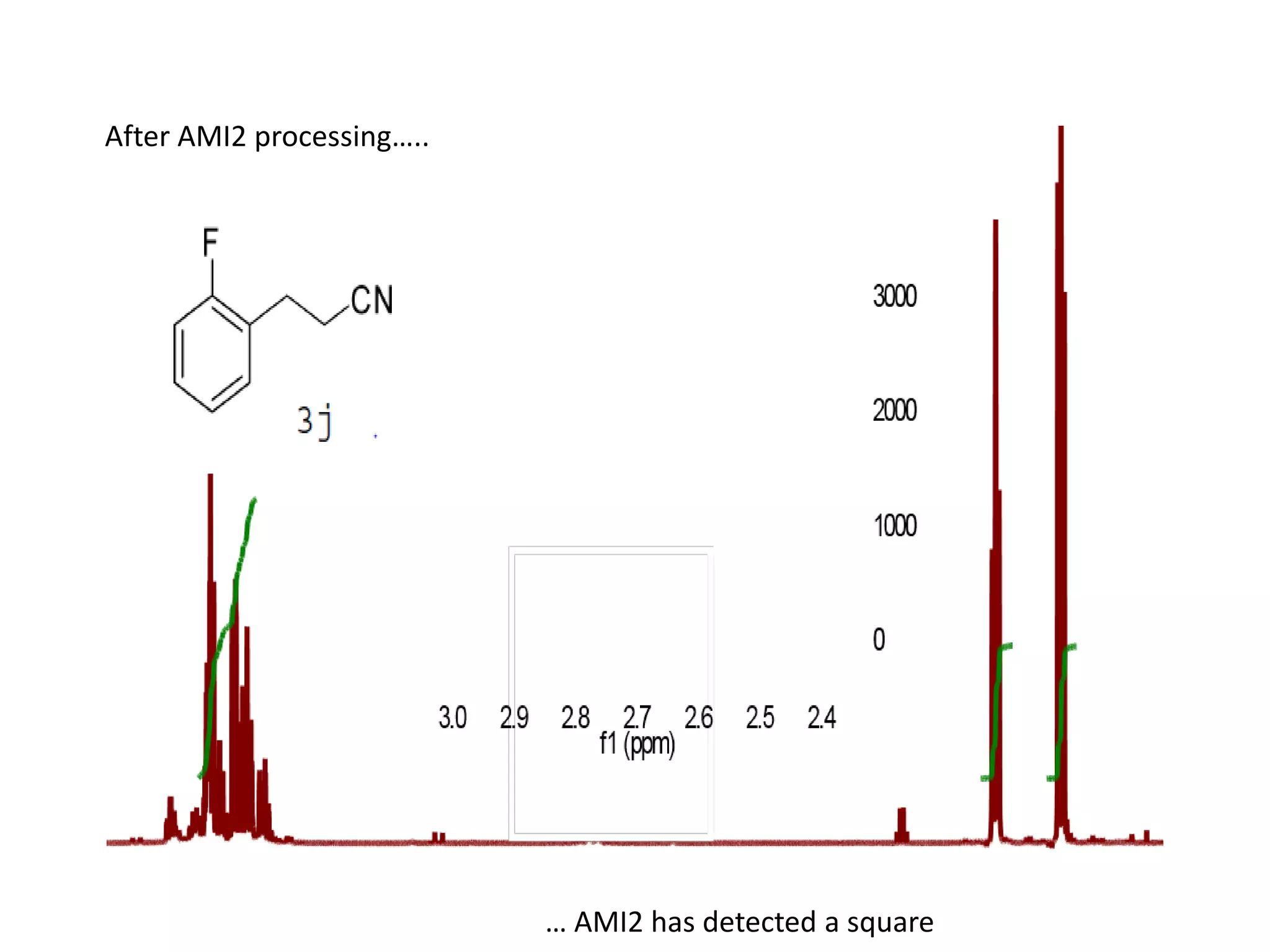
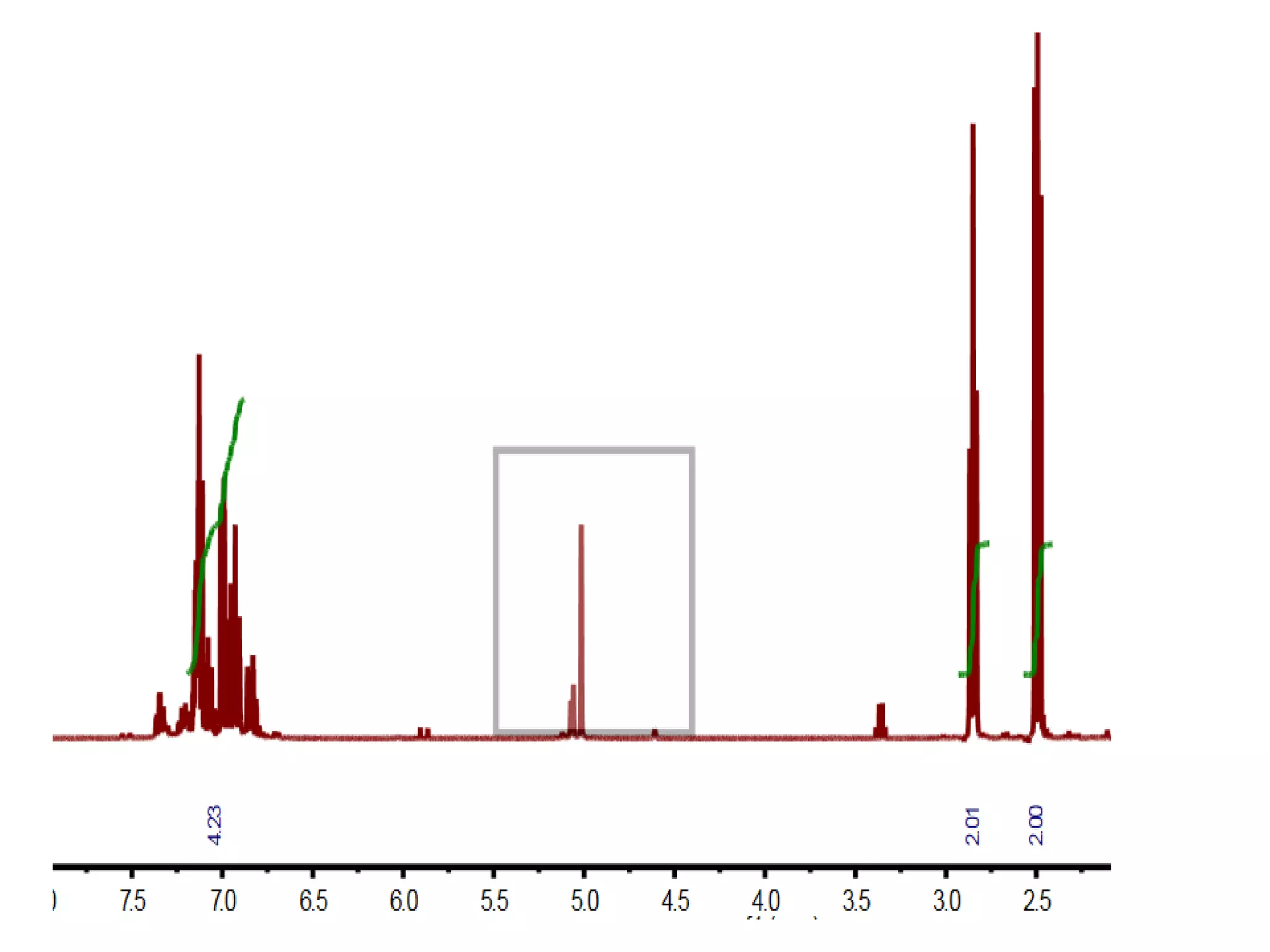
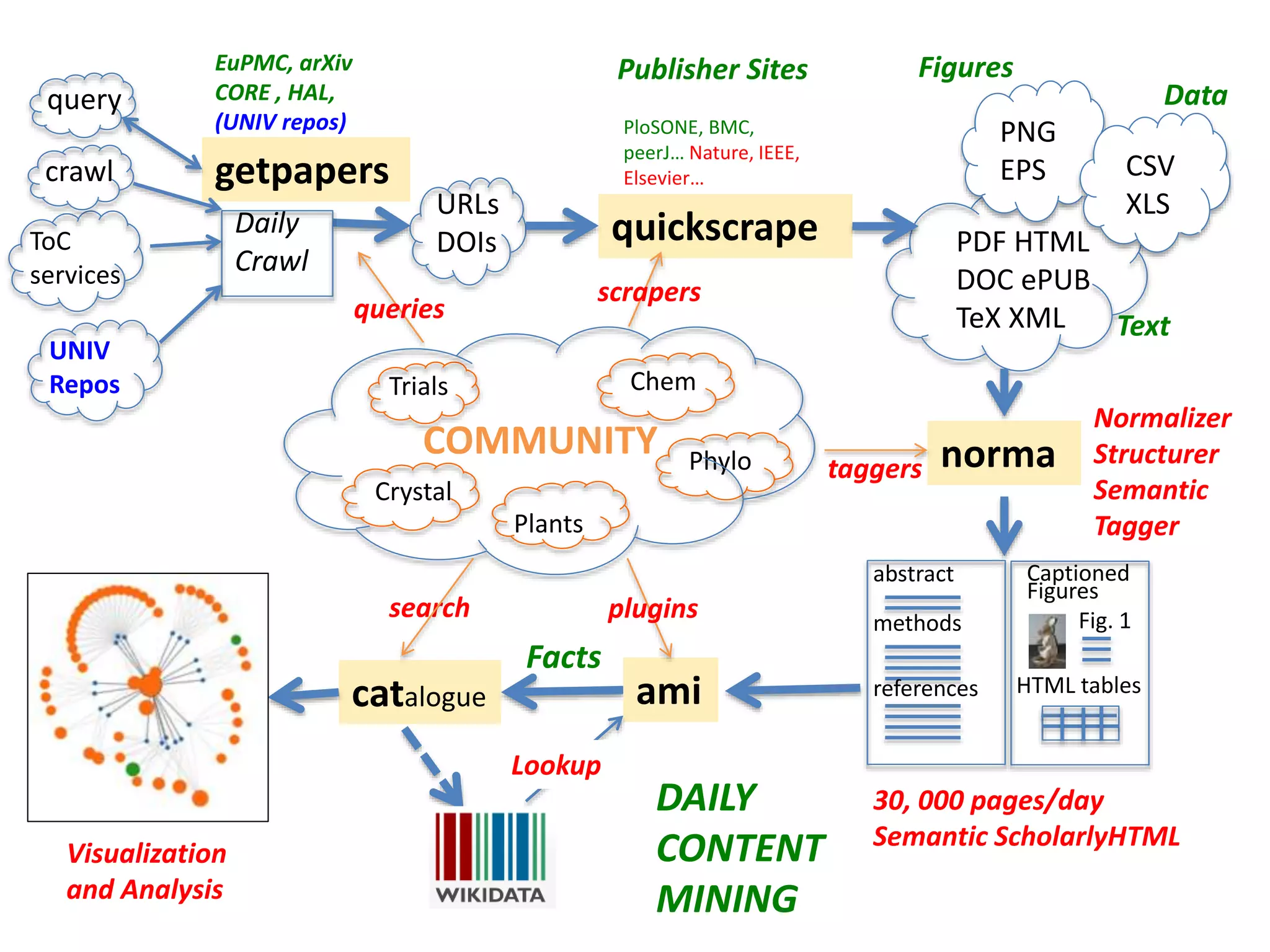
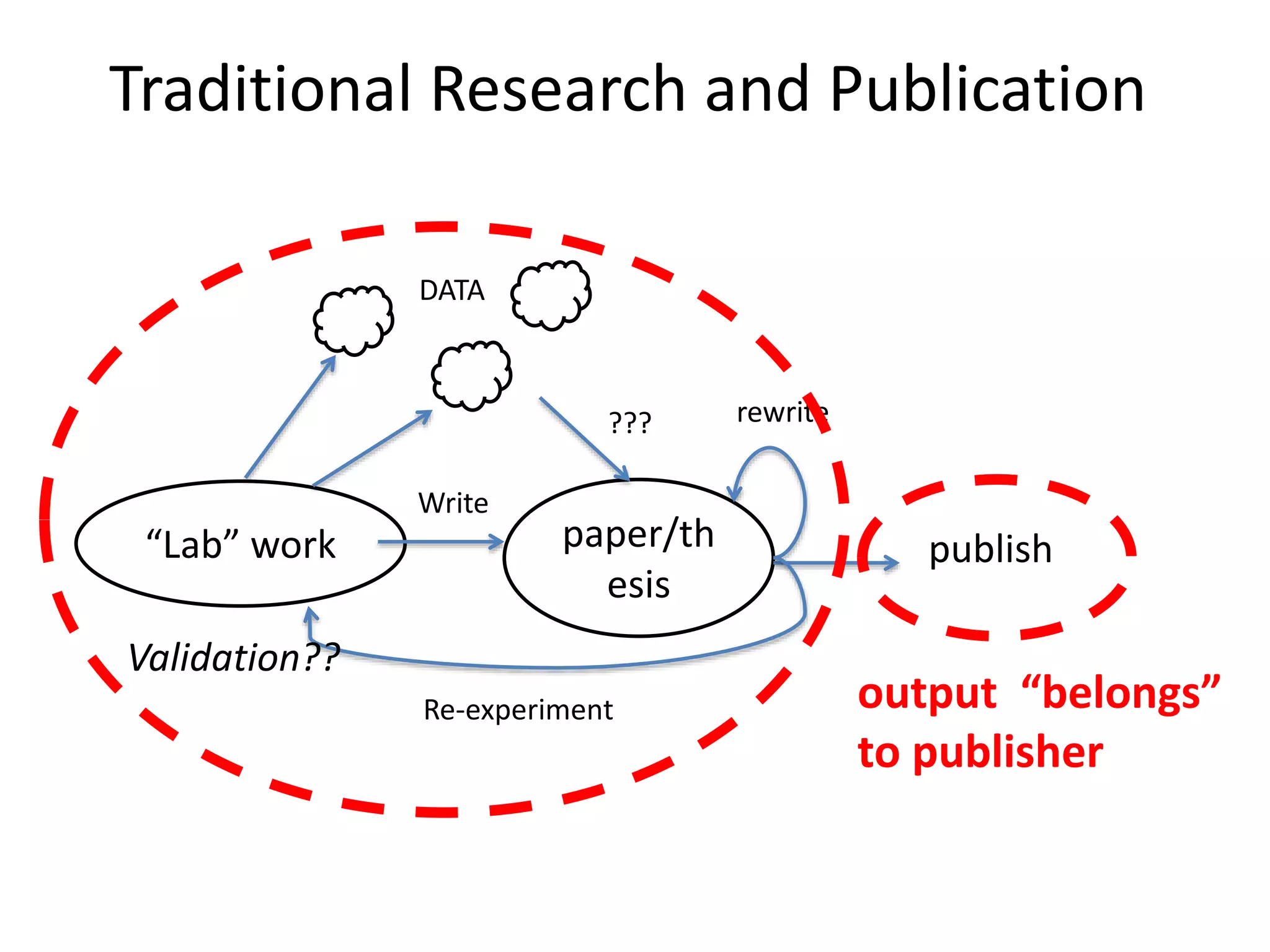
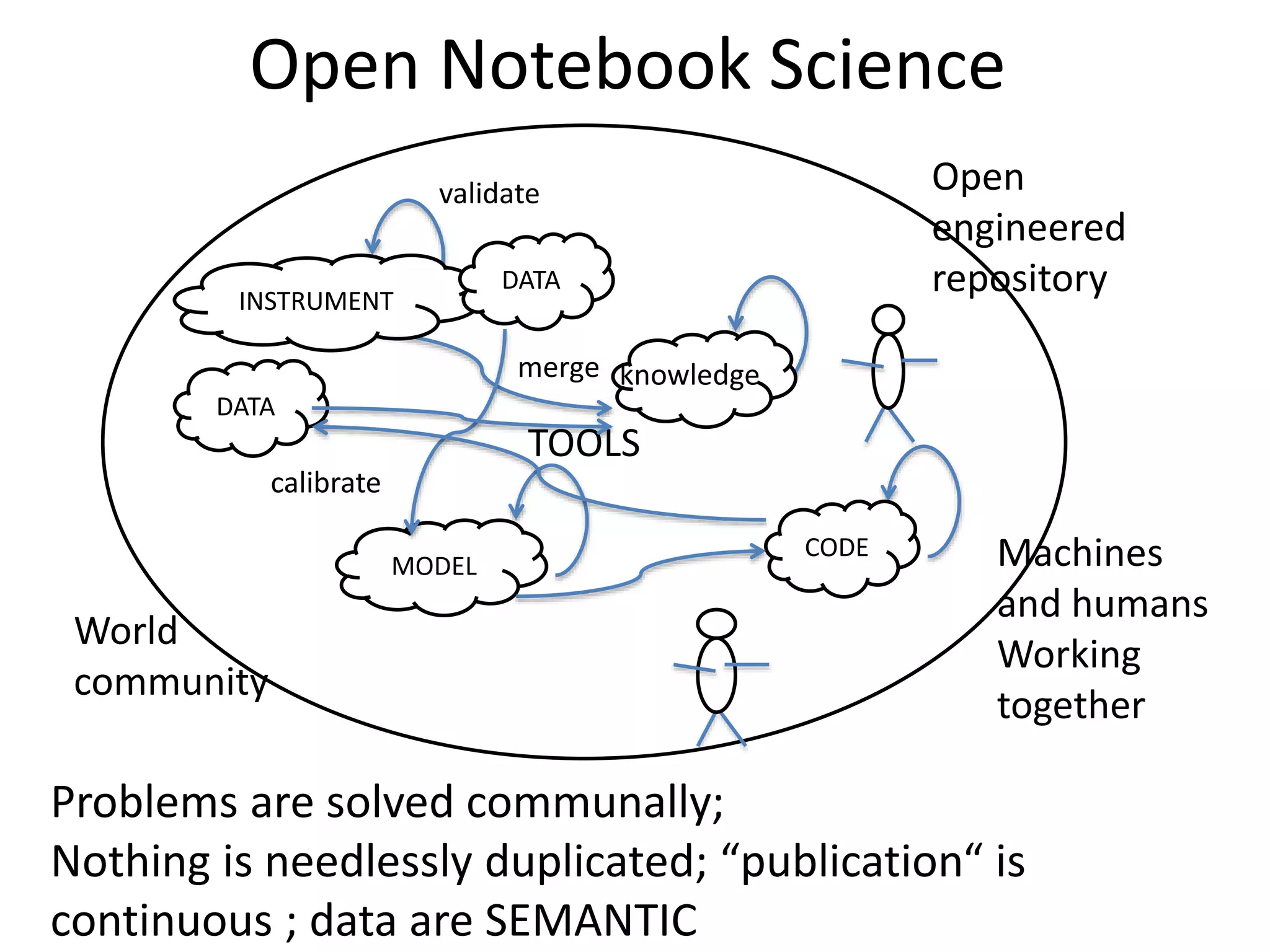
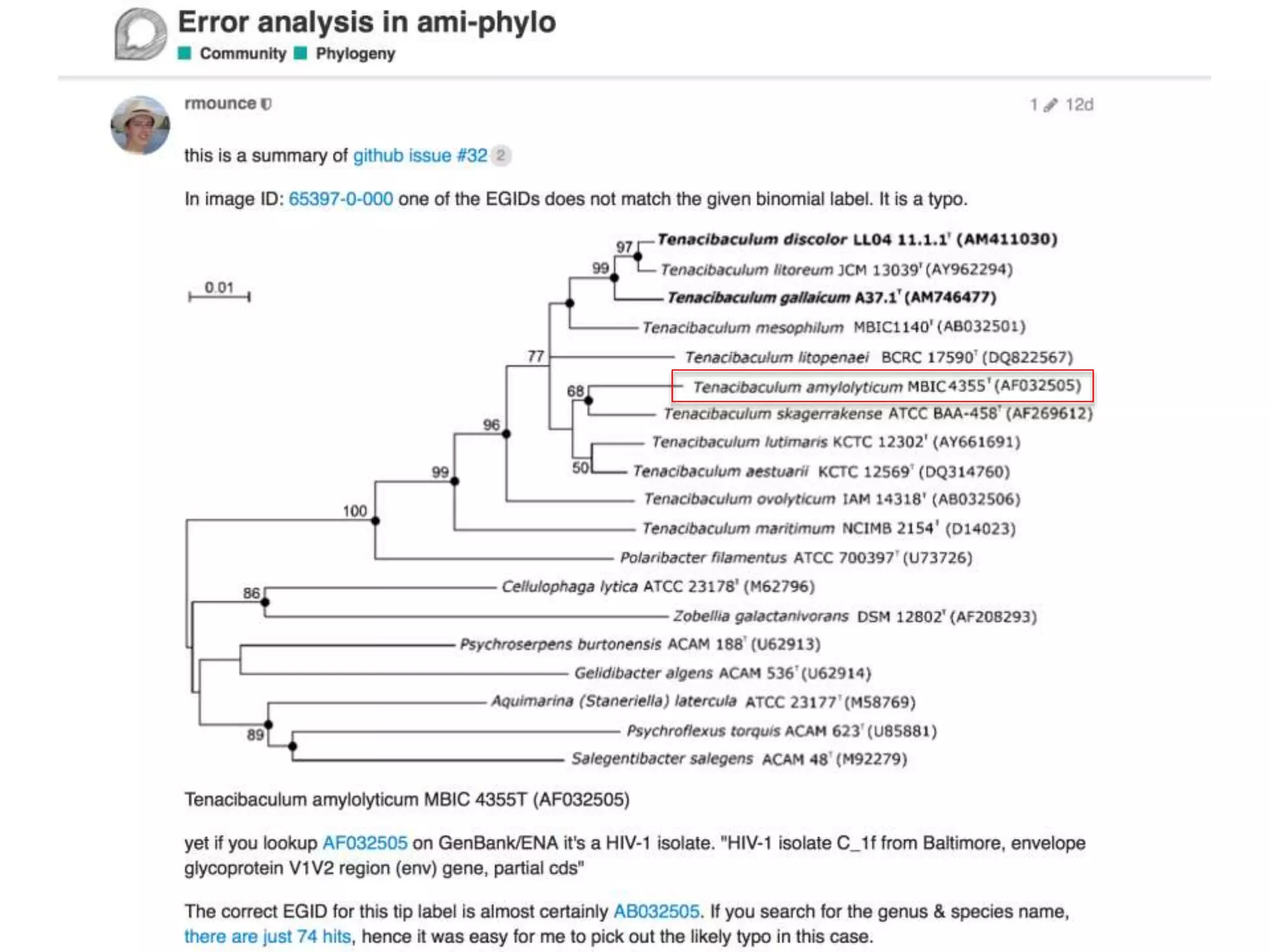
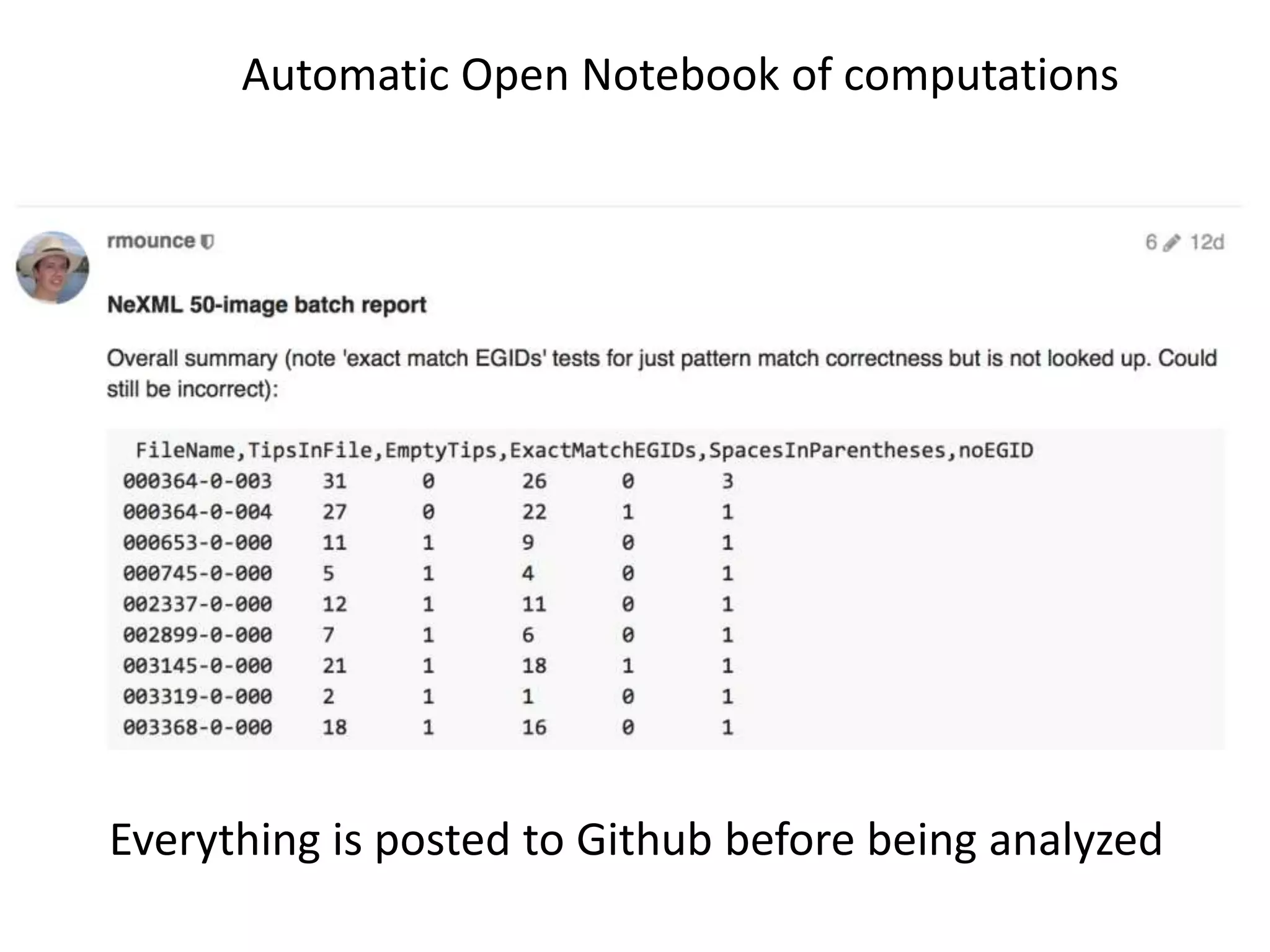
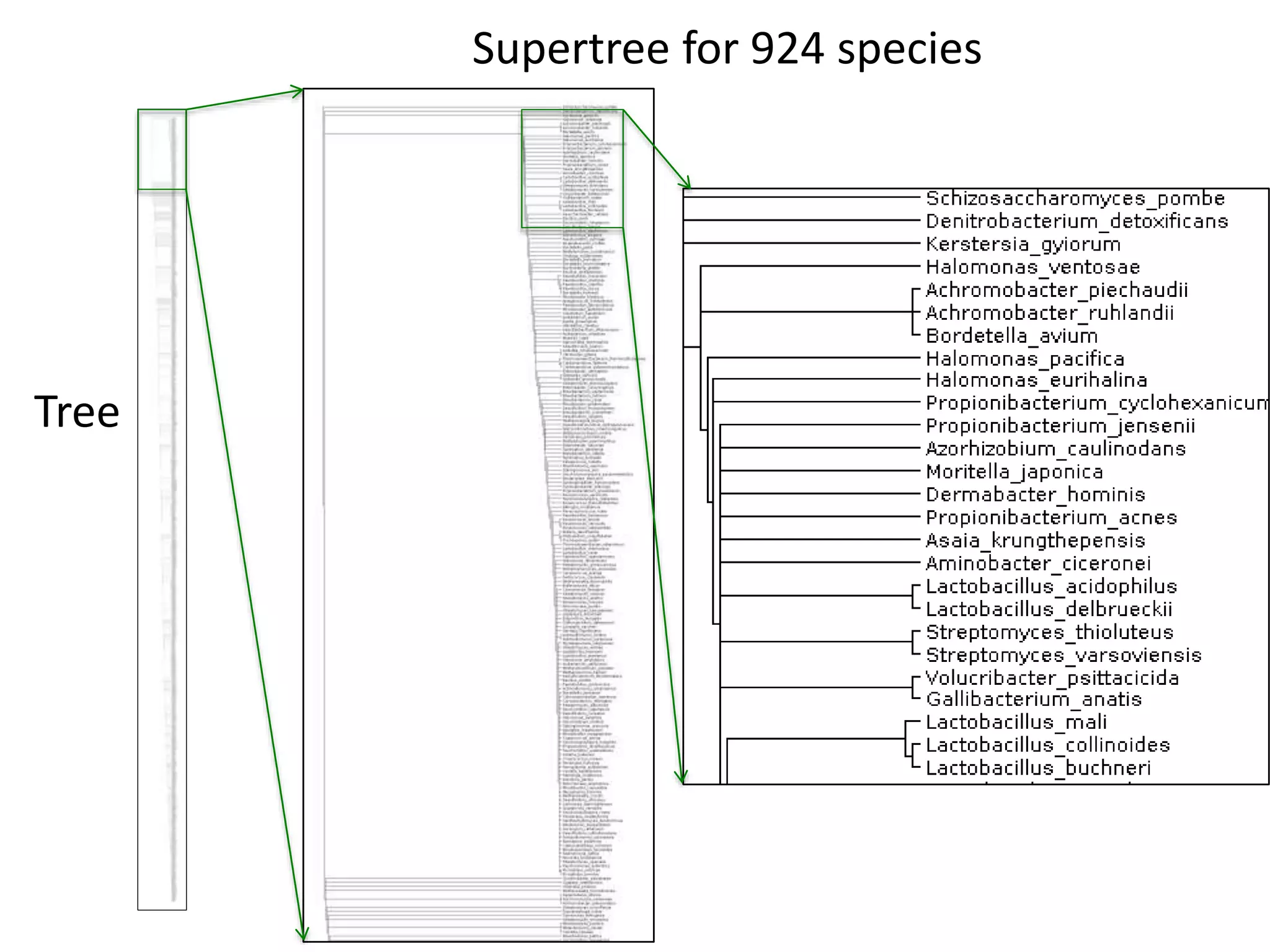
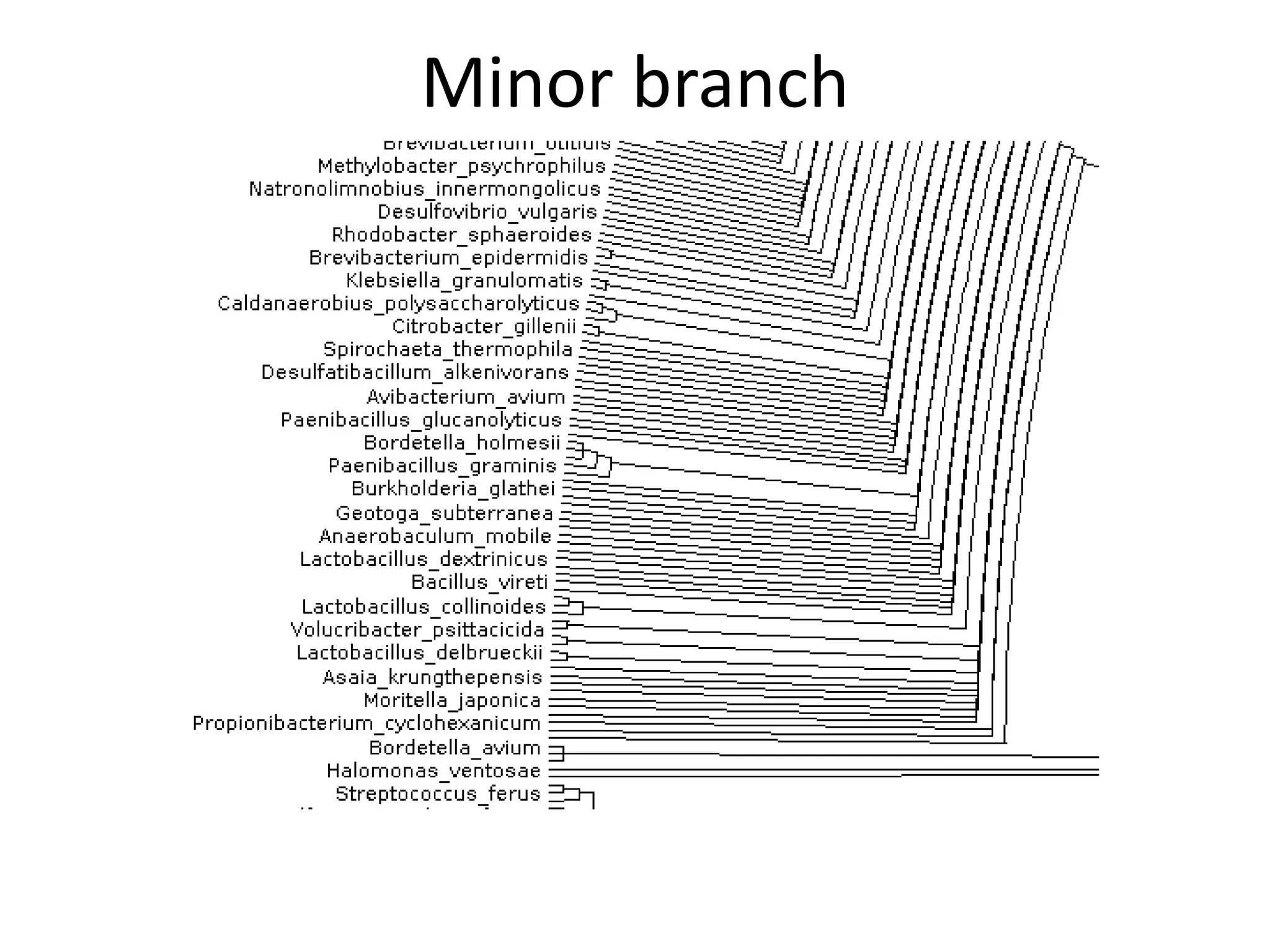
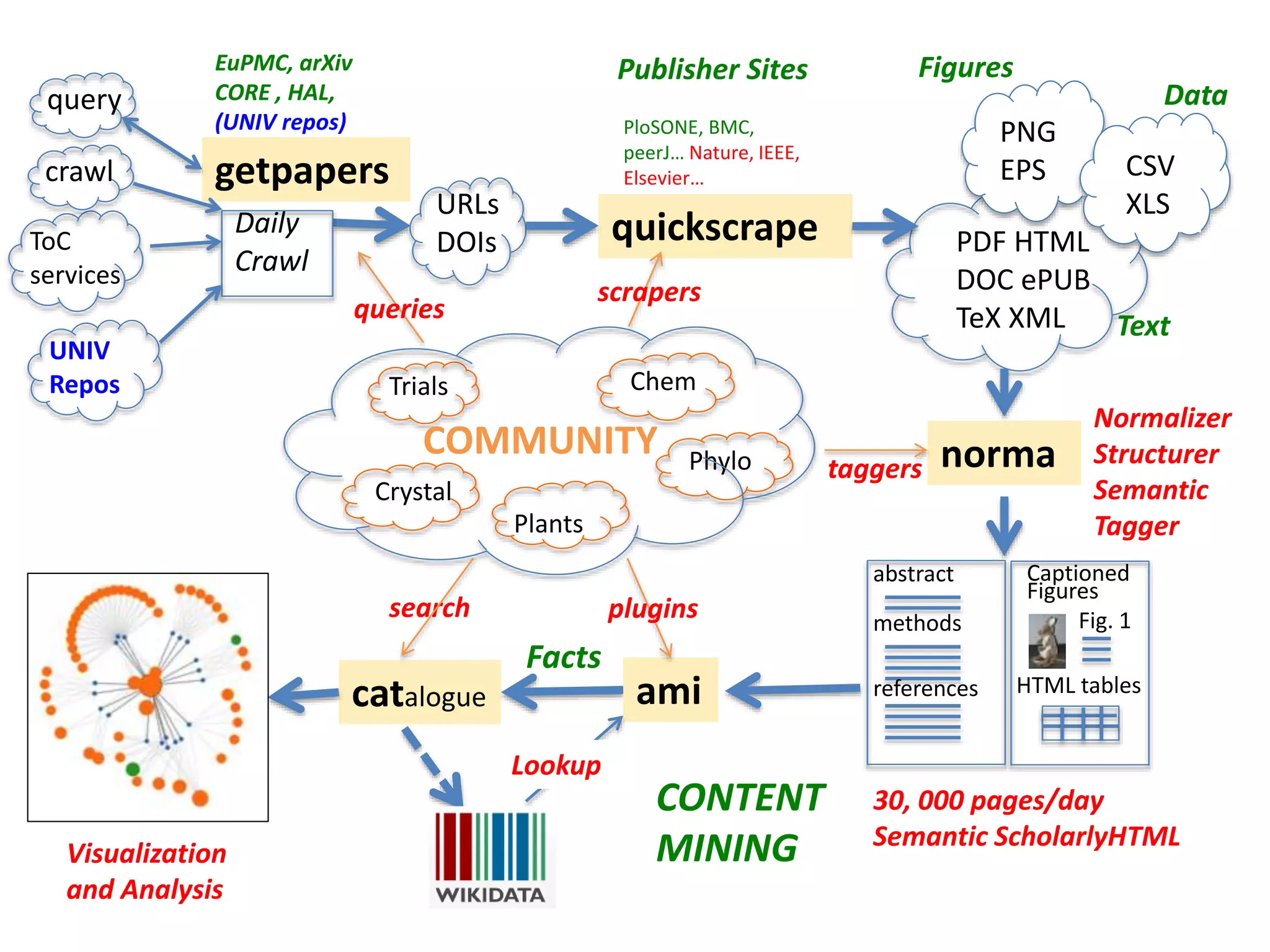
The document discusses the initiative ContentMine, which aims to liberate data hidden in bioscience literature through open technologies. It highlights the challenges of traditional scholarly publishing, including access restrictions and inefficiencies, while advocating for open science practices to improve data sharing and collaboration. The text mentions various projects and individuals involved in this movement and emphasizes the importance of enabling widespread access to scientific knowledge.






![http://www.nytimes.com/2015/04/08/opinion/yes-we-were-warned-about-
ebola.html
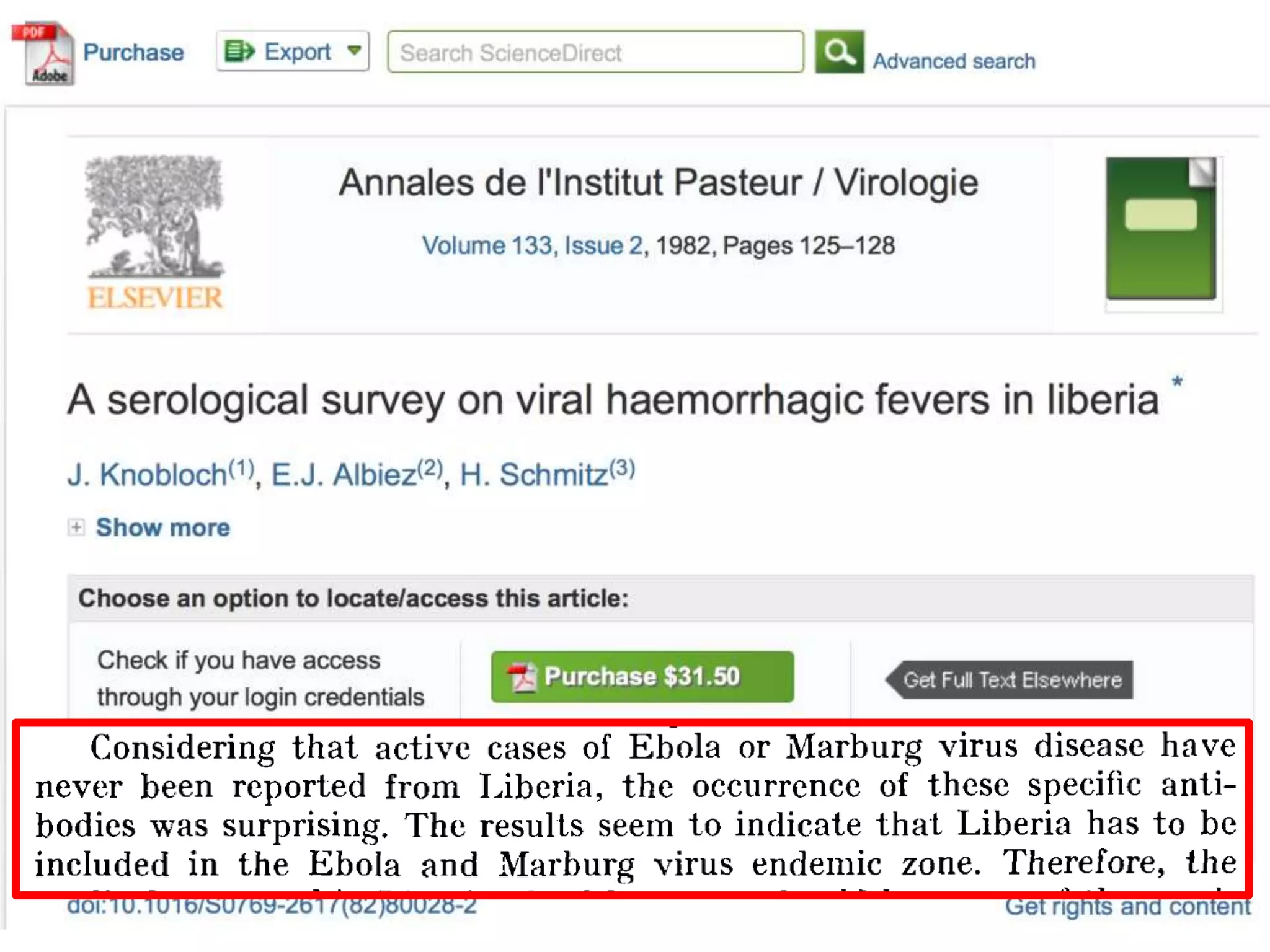
[Liberian Ministry of Health] were stunned recently when we stumbled across
an article by European researchers in Annals of Virology [1982]: “The results
seem to indicate that Liberia has to be included in the Ebola virus endemic
zone.” In the future, the authors asserted, “medical personnel in Liberian health
centers should be aware of the possibility that they may come across active
cases and thus be prepared to avoid nosocomial epidemics,” referring to
hospital-acquired infection.
Adage in public health: “The road to inaction is paved with research
papers.”
Bernice Dahn (chief medical officer of Liberia’s Ministry of Health)
Vera Mussah (director of county health services)
Cameron Nutt (Ebola response adviser to Partners in Health)
A System Failure of Scholarly Publishing](https://image.slidesharecdn.com/neuroscience2015a-151022054611-lva1-app6892-151124124652-lva1-app6891/75/ContentMining-in-Neuroscience-10-2048.jpg)



![Output of scholarly publishing
[2] https://en.wikipedia.org/wiki/Mont_Blanc#/media/File:Mont_Blanc_depuis_Valmorel.jpg
586,364 Crossref DOIs 201,507 [1] per month
1.5 million (papers + supplemental data) /year [citation needed]*
each 3 mm thick
4500 m high per year [2]
* Most is not Publicly readable
[1] http://www.crossref.org/01company/crossref_indicators.html](https://image.slidesharecdn.com/neuroscience2015a-151022054611-lva1-app6892-151124124652-lva1-app6891/75/ContentMining-in-Neuroscience-16-2048.jpg)
![Scientific and Medical publication (STM)[+]
• World Citizens pay $450,000,000,000…
• … for research in 1,500,000 articles …
• … cost $300,000 each to create …
• … $7000 each to “publish” [*]…
• … $10,000,000,000 from academic libraries …
• … to “publishers” who forbid access to 99.9% of citizens of
the world …
• 85% of medical research is wasted (not published, badly
conceived, duplicated, …) [Lancet 2009]
[+] Figures probably +- 50 %
[*] arXiV preprint server costs $7 USD per paper](https://image.slidesharecdn.com/neuroscience2015a-151022054611-lva1-app6892-151124124652-lva1-app6891/75/ContentMining-in-Neuroscience-17-2048.jpg)









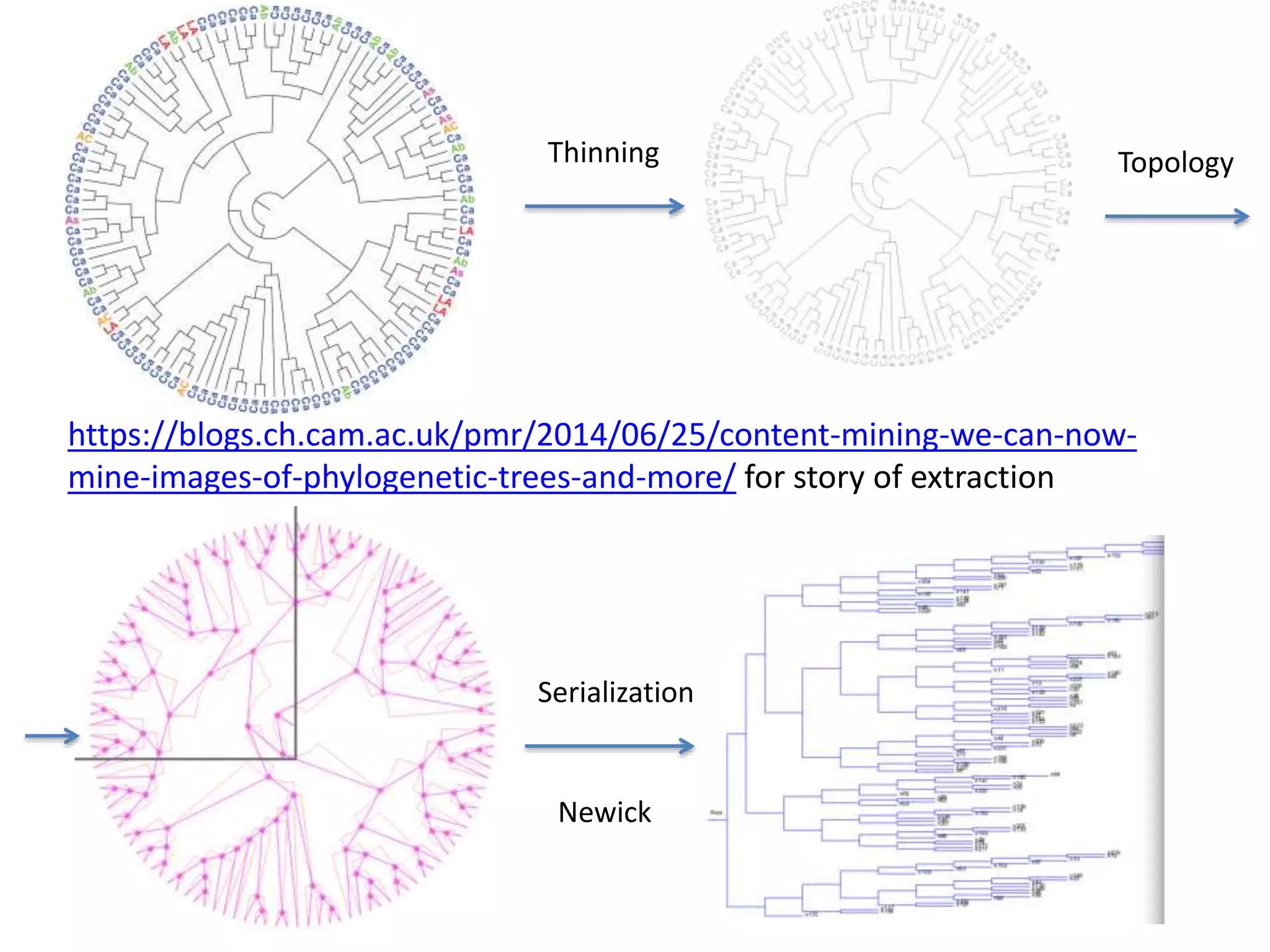

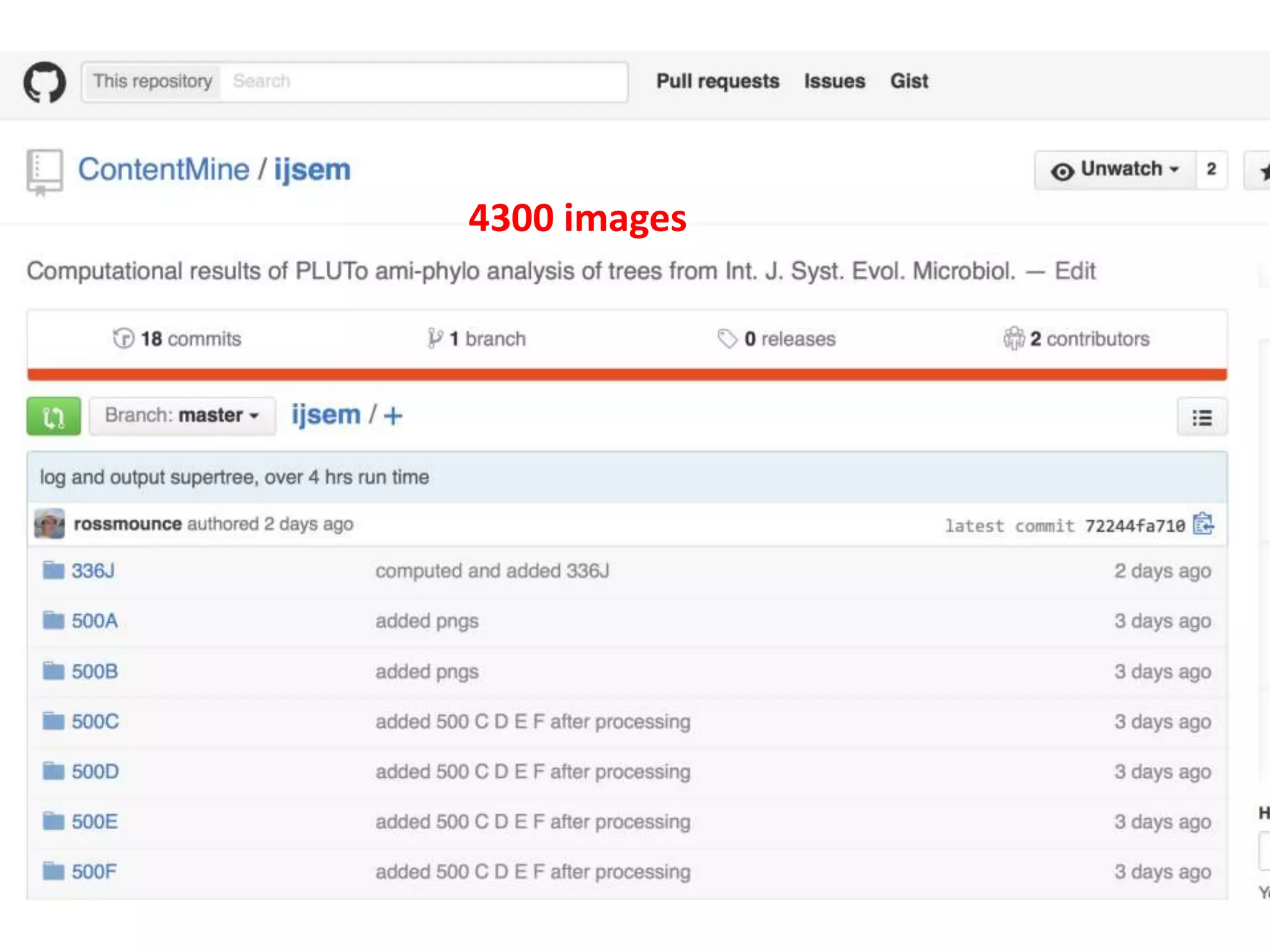
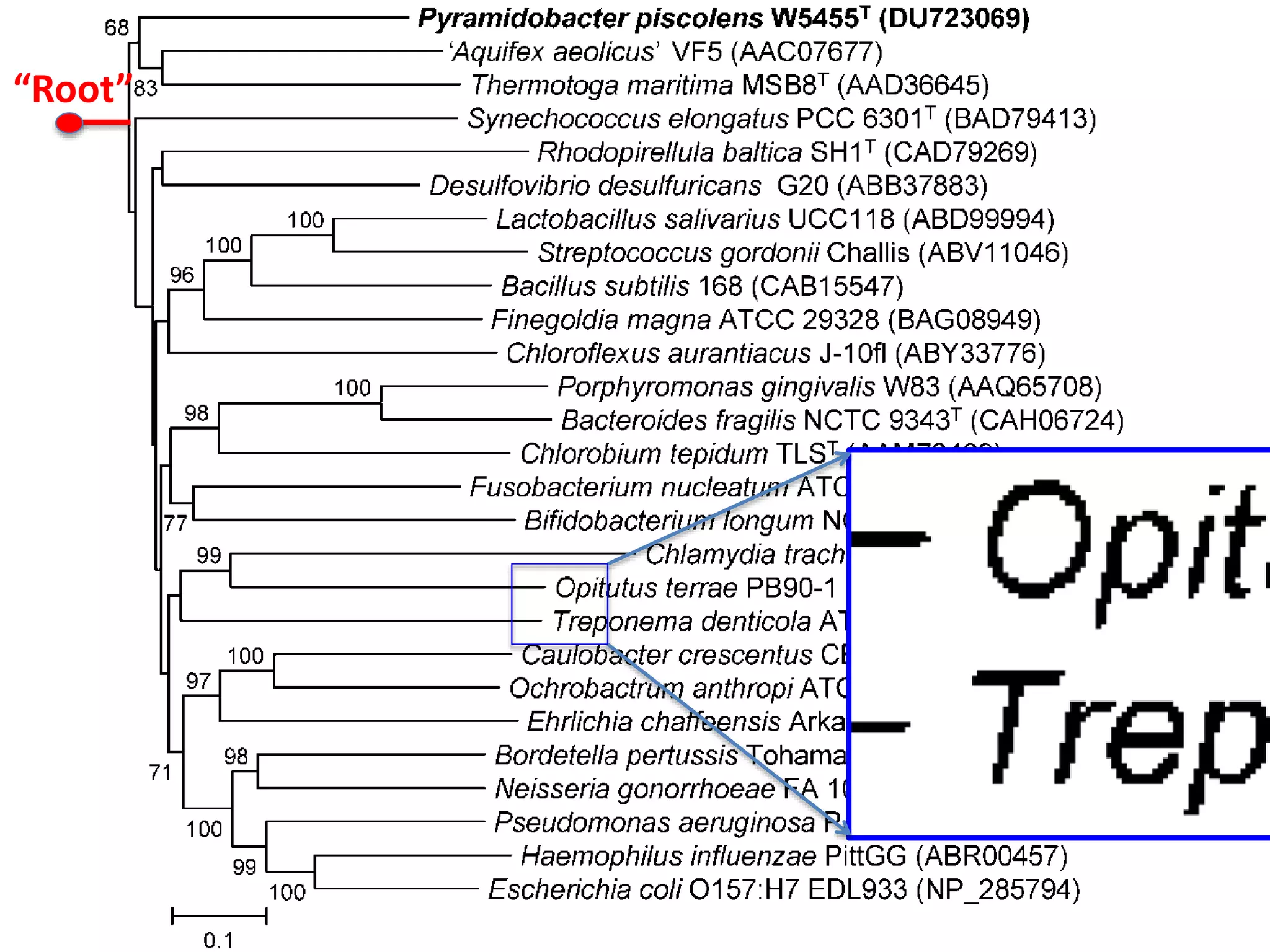
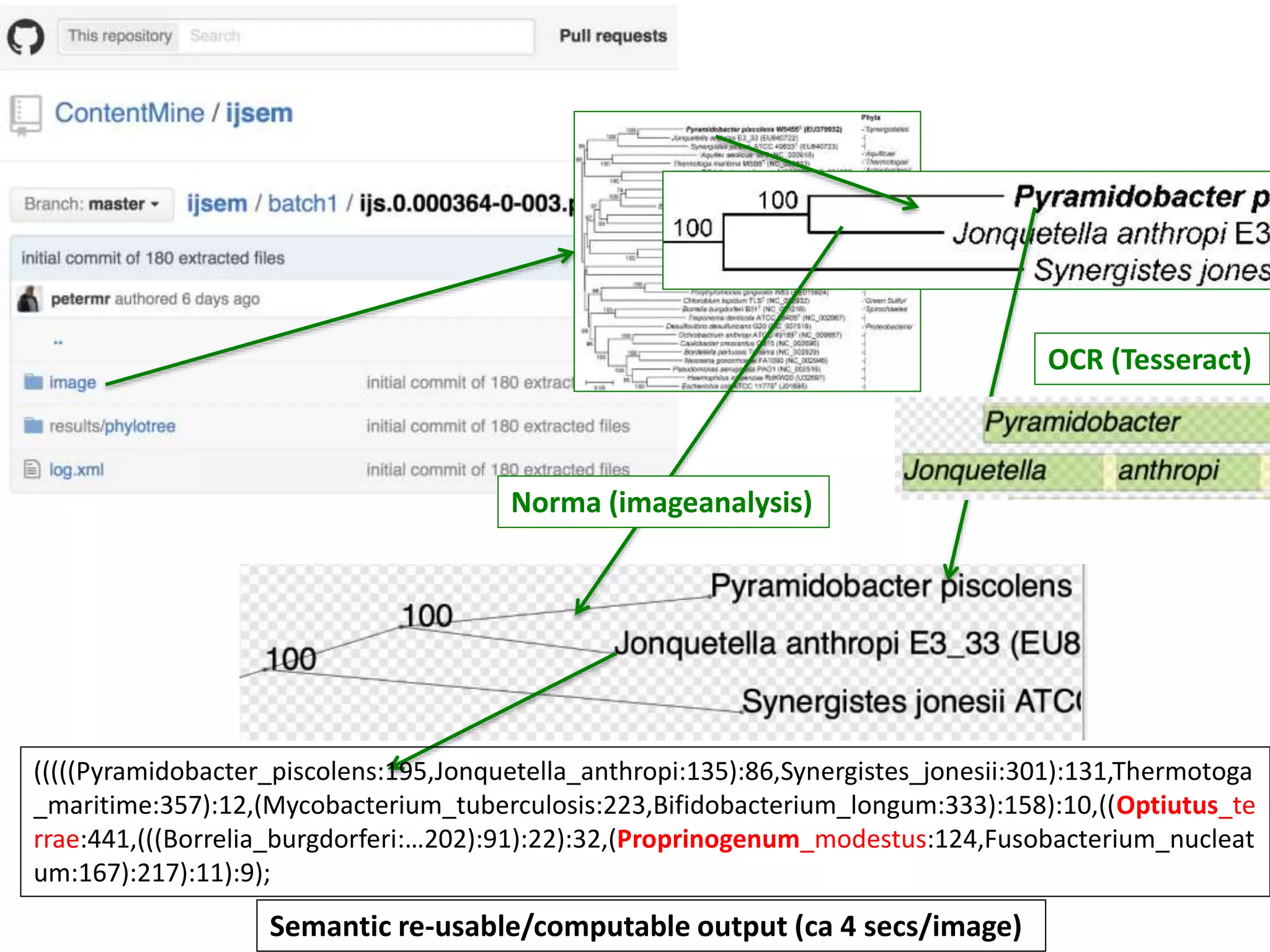
![Bacillus subtilis [131238]*
Bacteroides fragilis [221817]
Brevibacillus brevis
Cyclobacterium marinum
Escherichia coli [25419]
Filobacillus milosensis
Flectobacillus major [15809775]
Flexibacter flexilis [15809789]
Formosa algae
Gelidibacter algens [16982233]
Halobacillus halophilus
Lentibacillus salicampi [18345921]
Octadecabacter arcticus
Psychroflexus torquis [16988834]
Pseudomonas aeruginosa [31856]
Sagittula stellata [16992371]
Salegentibacter salegens
Sphingobacterium spiritivorum
Terrabacter tumescens
• [Identifier in Wikidata]
• Missing = not found with Wikidata API
20 commonest organisms (in > 30 papers) in trees from IJSEM*
Half do not appear to be in Wikidata
Can the Wikipedia Scientists comment?
*Int. J. Syst. Evol. Microbiol.](https://image.slidesharecdn.com/neuroscience2015a-151022054611-lva1-app6892-151124124652-lva1-app6891/75/ContentMining-in-Neuroscience-51-2048.jpg)


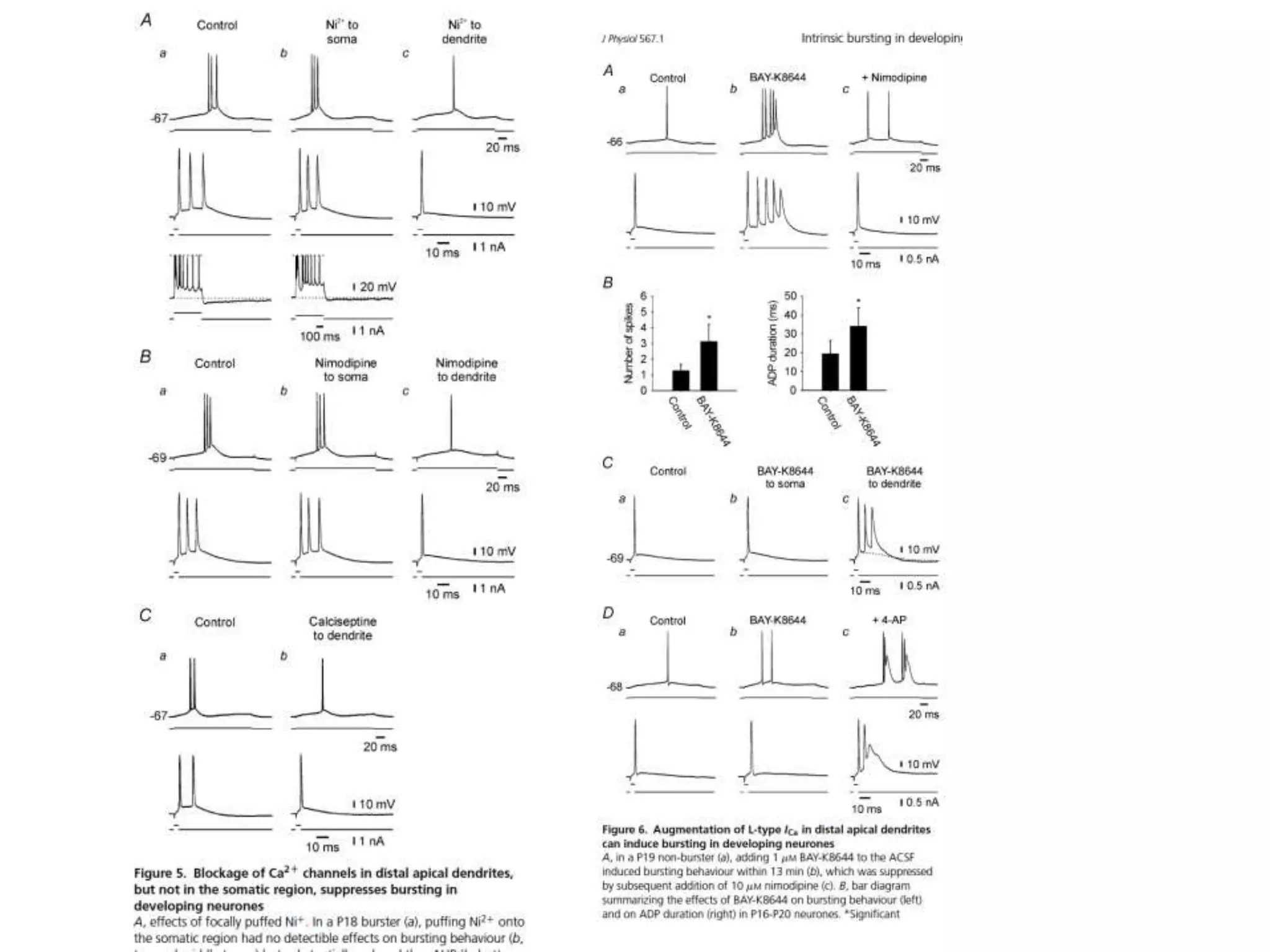
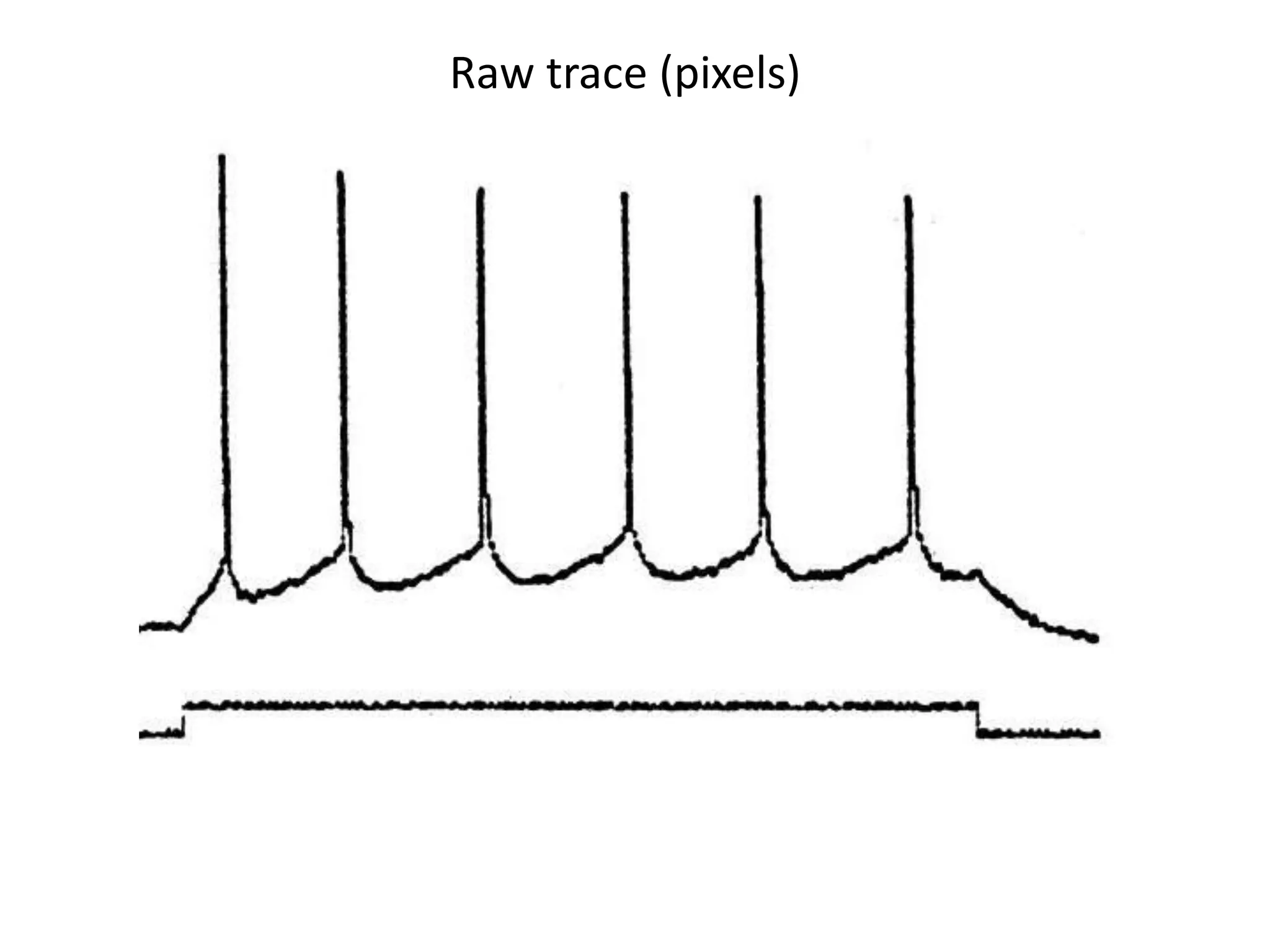
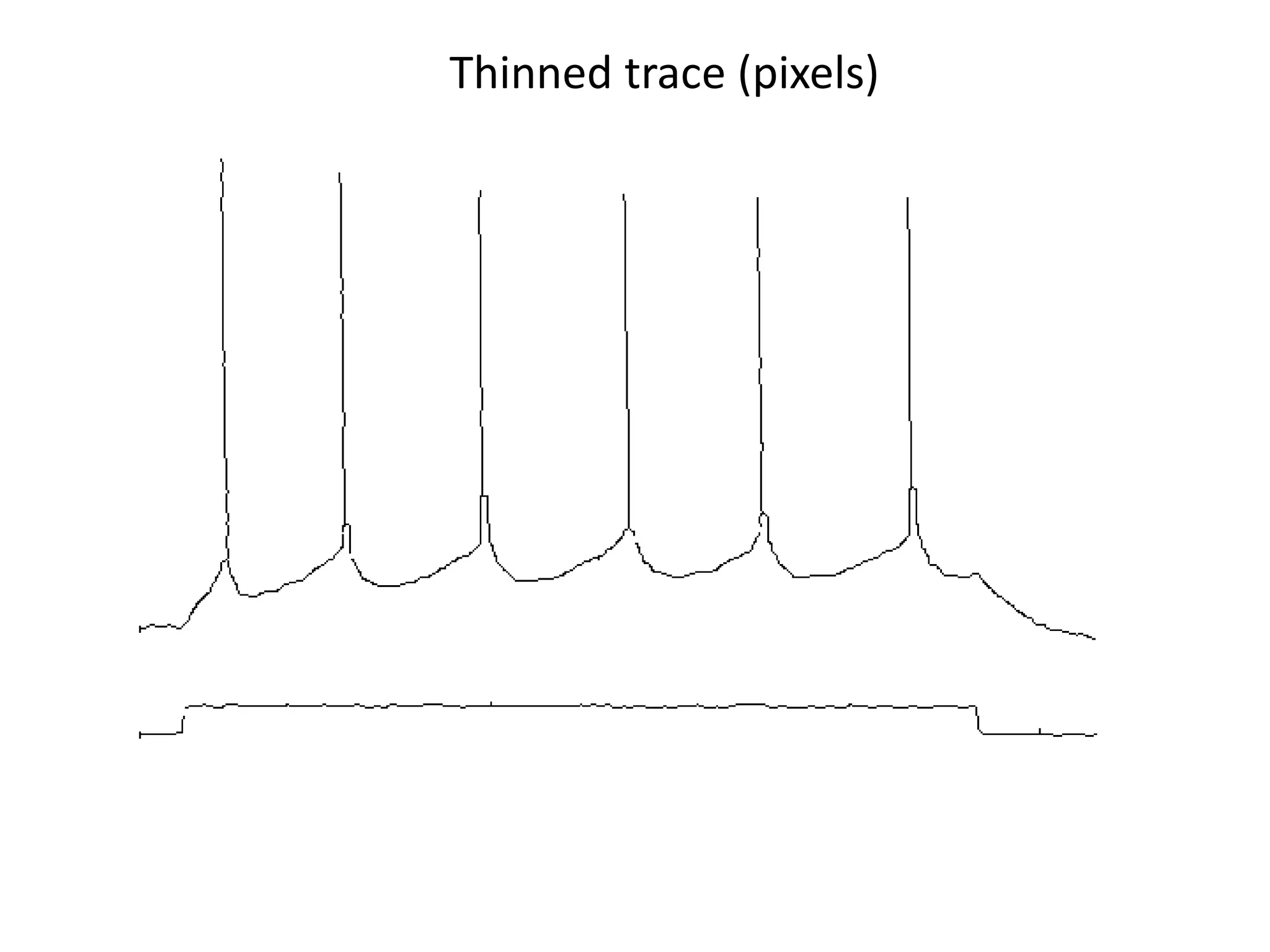
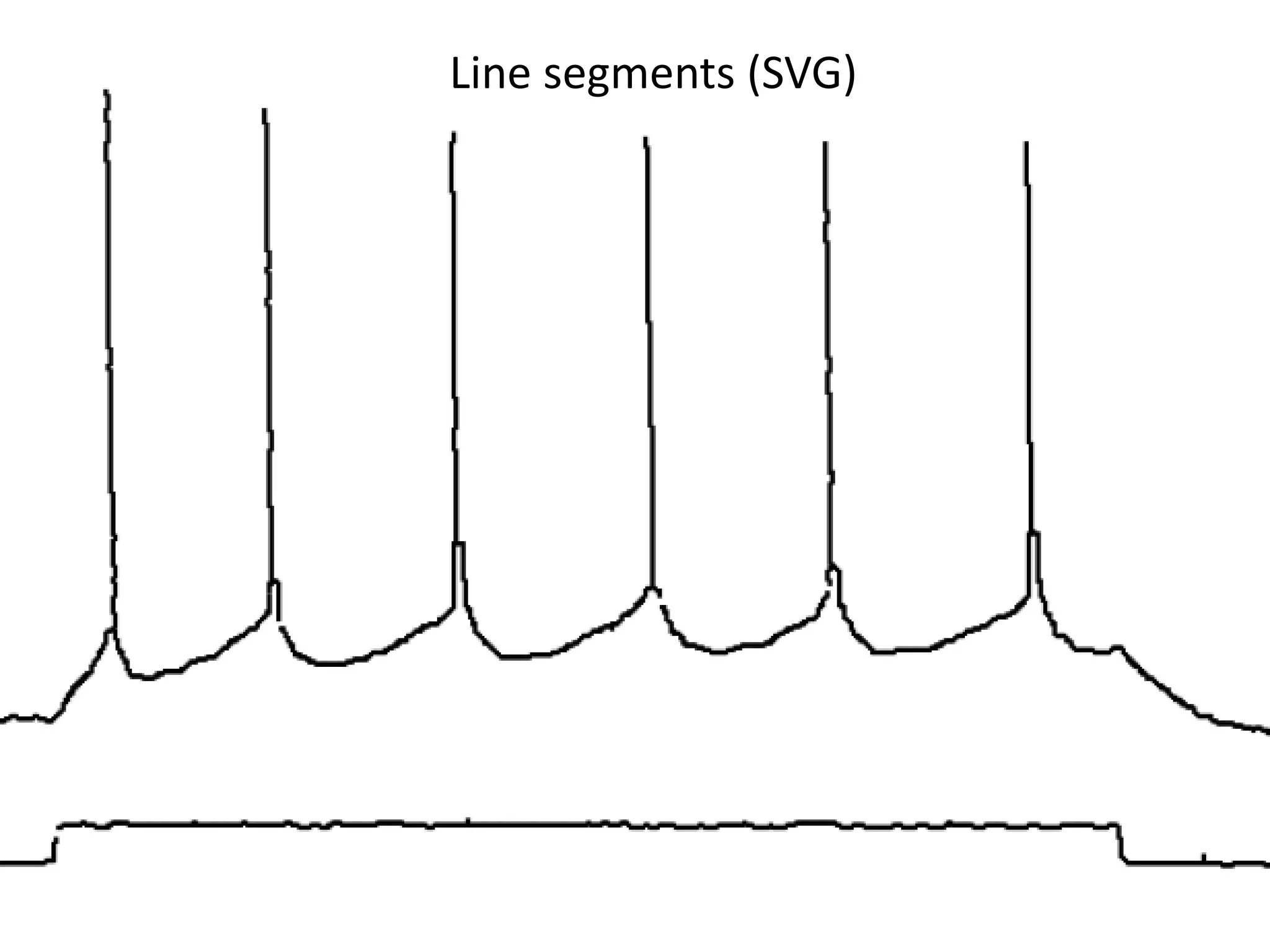

![Elsevier wants to control Open Data
[asked by Michelle Brook]](https://image.slidesharecdn.com/neuroscience2015a-151022054611-lva1-app6892-151124124652-lva1-app6891/75/ContentMining-in-Neuroscience-71-2048.jpg)