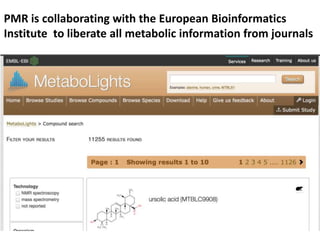
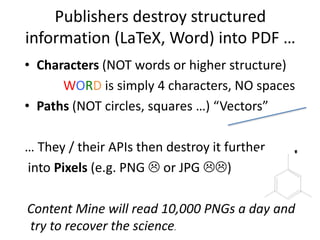
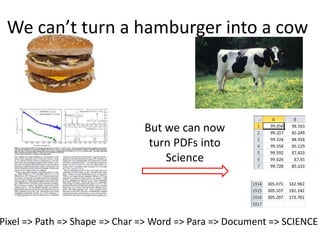
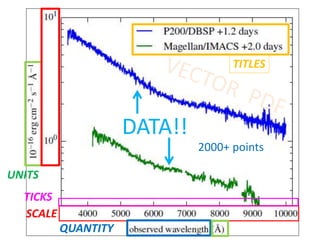
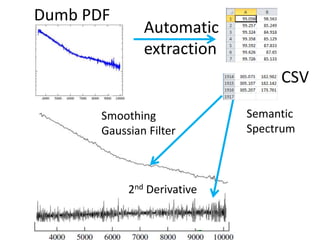
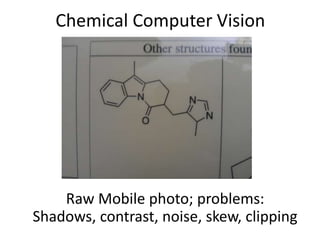
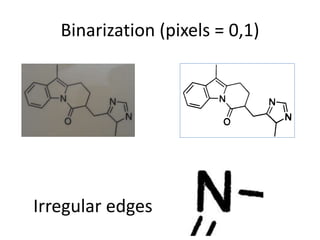
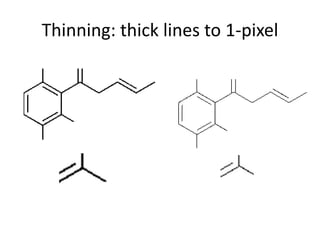
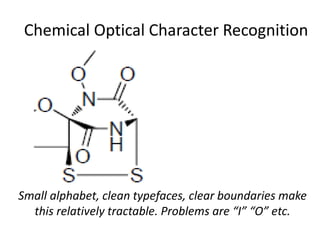
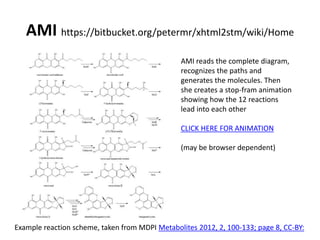
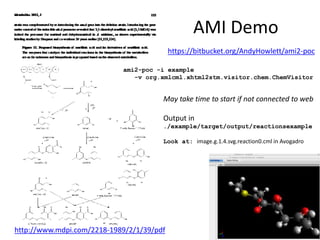
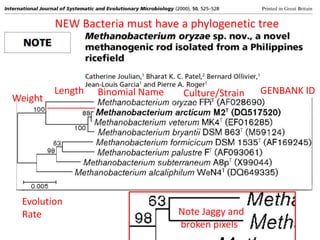
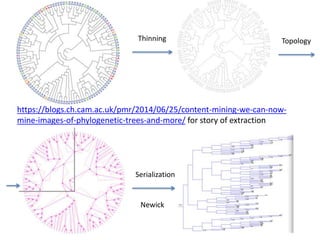
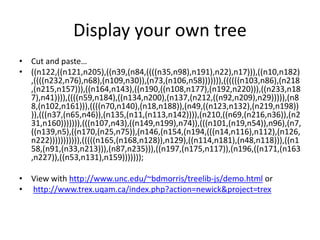
The document discusses the ContentMine project, which aims to mine and extract information from scientific literature, particularly focusing on phylogenetic trees and chemical data. It highlights the challenges of working with various formats, especially how publishers convert structured information into less accessible formats like PDF and images. The project employs advanced techniques for data extraction and processing to liberate and utilize scientific data effectively.