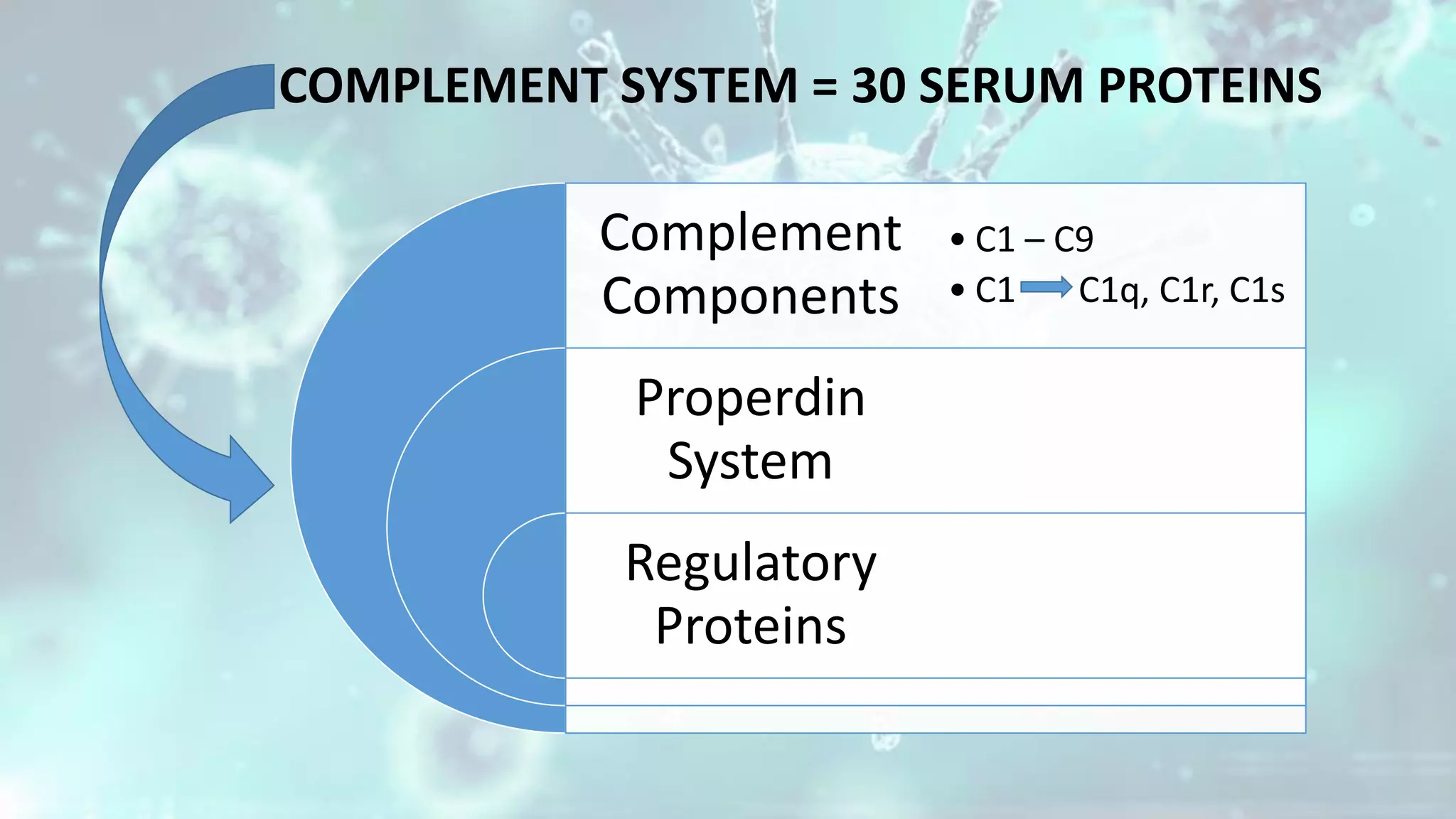
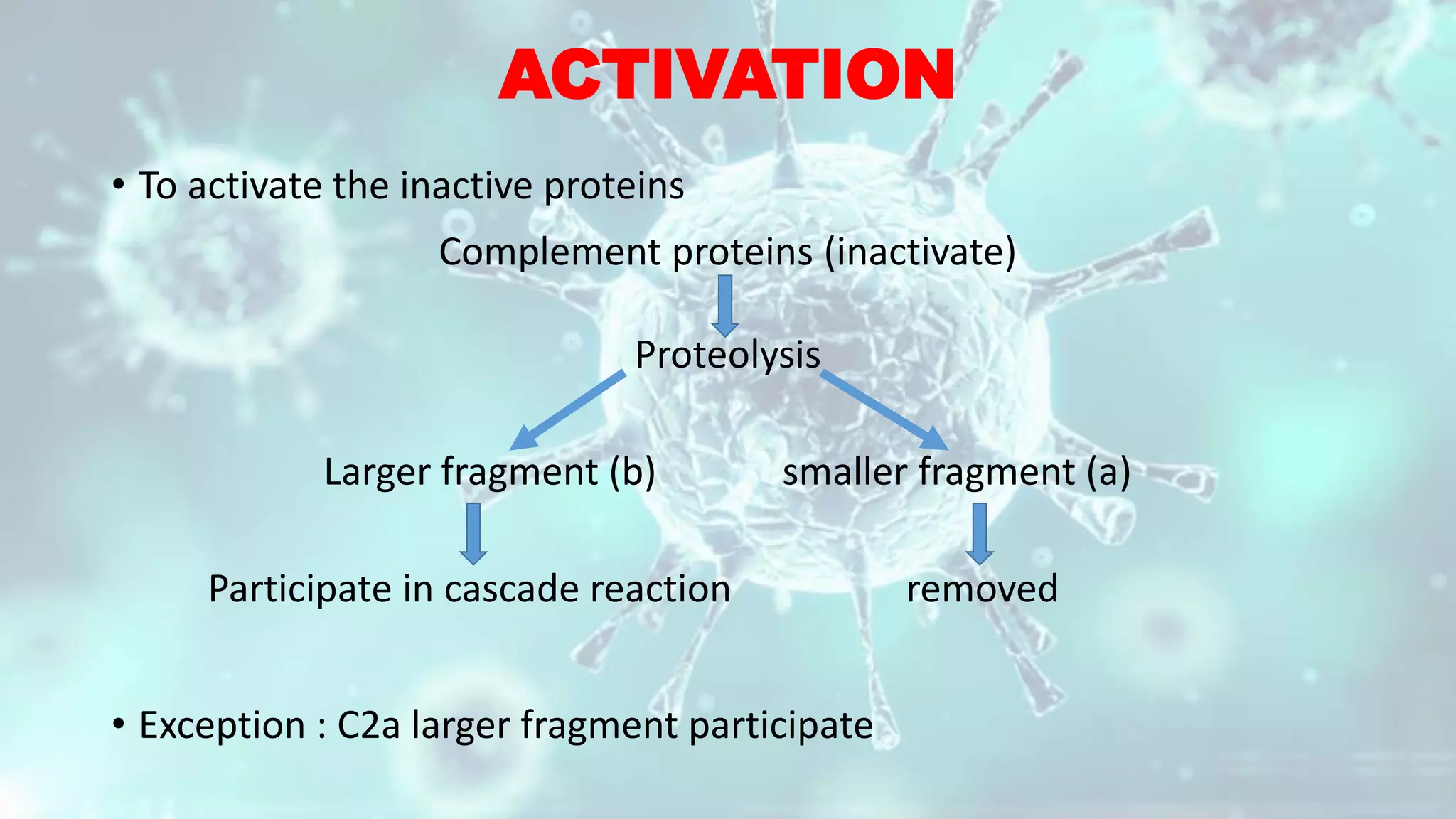
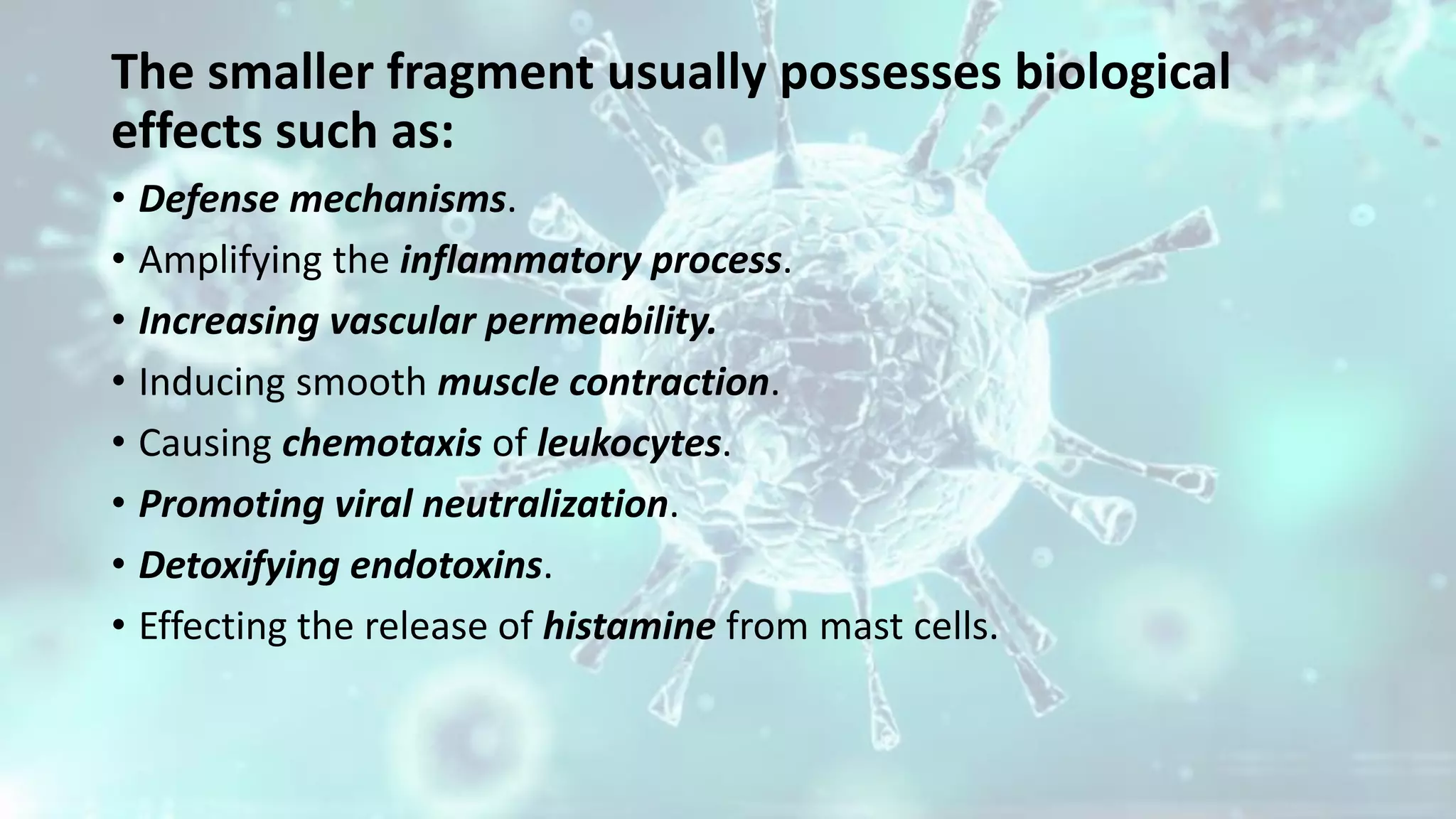
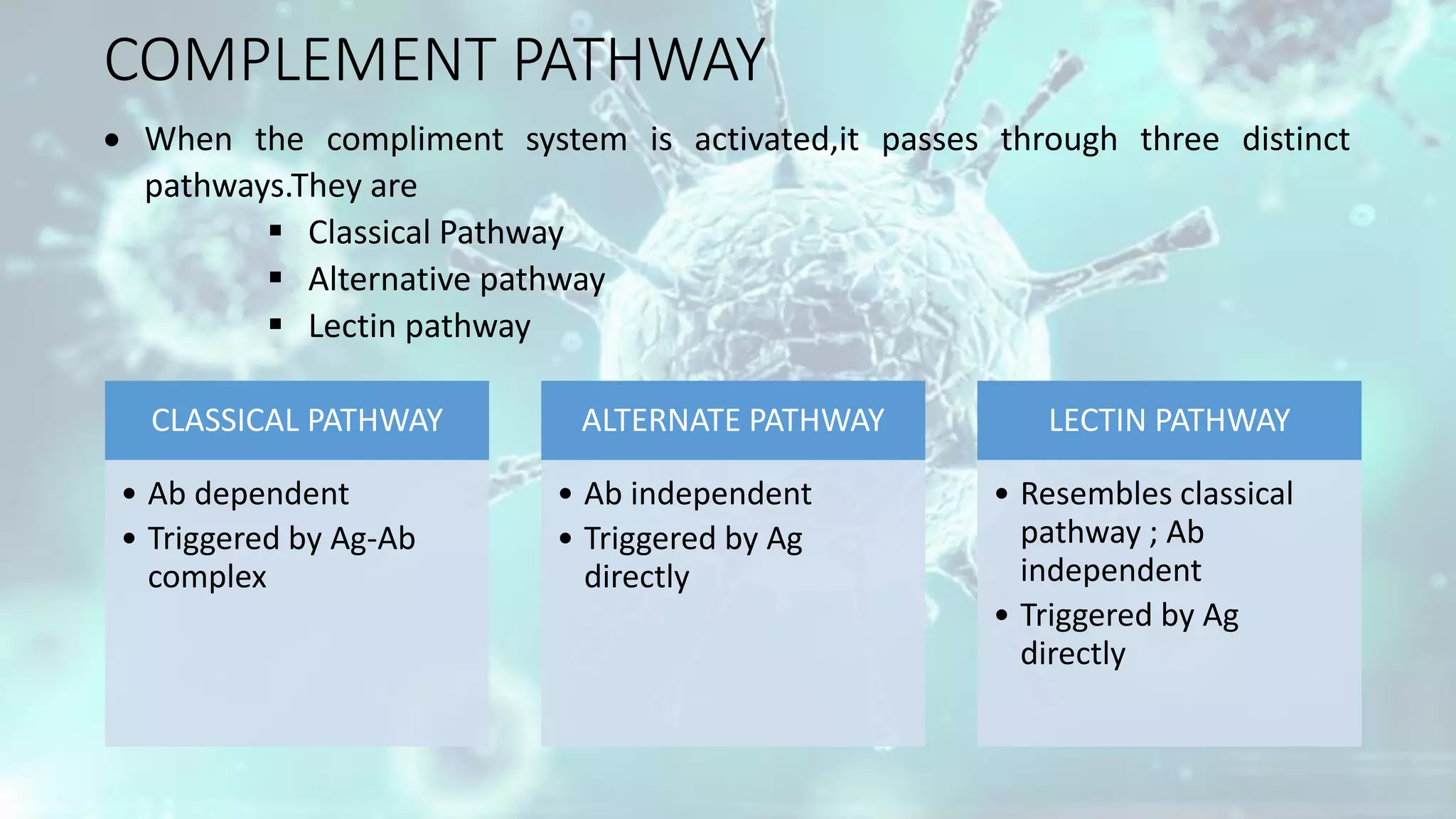
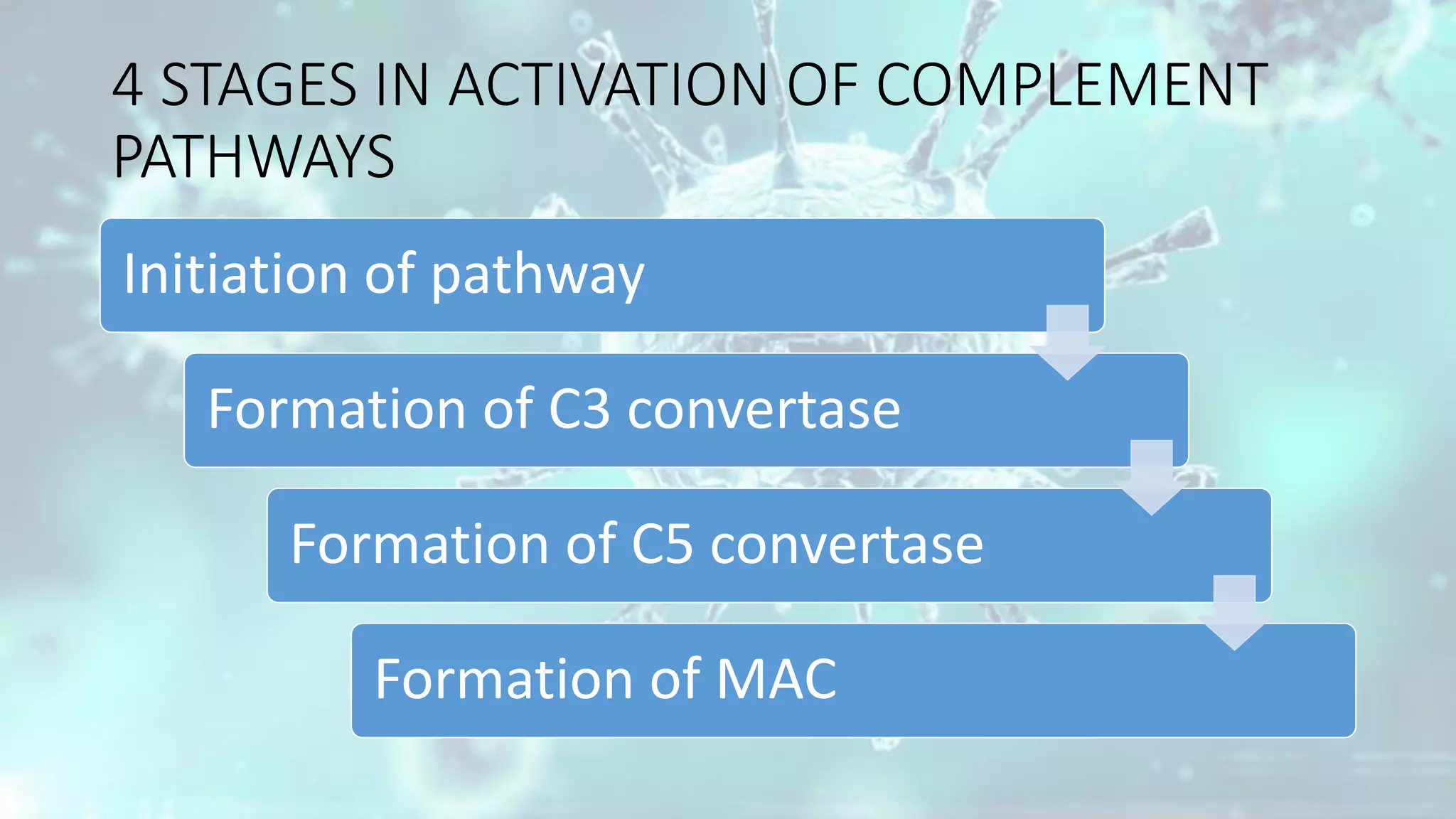
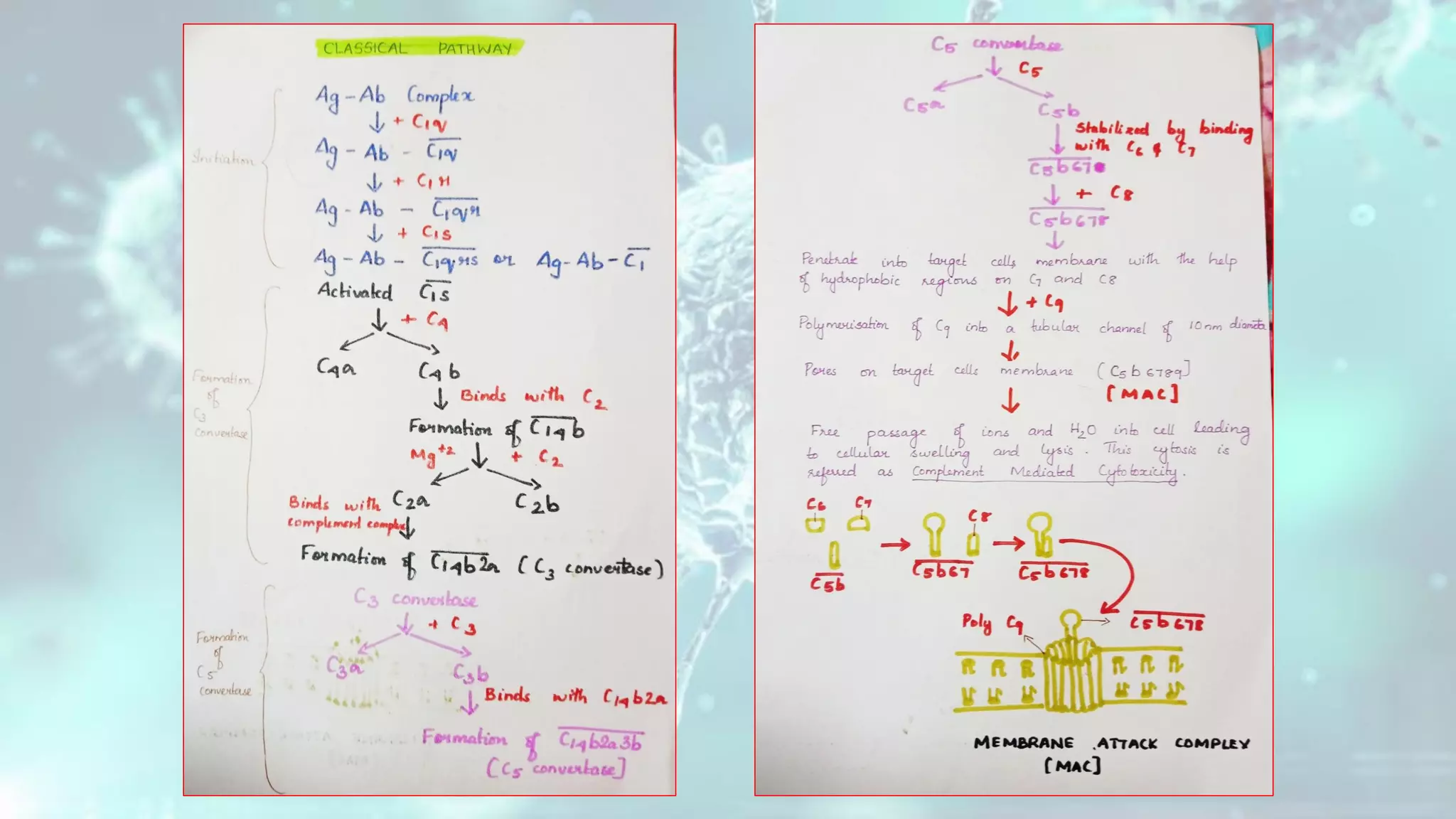
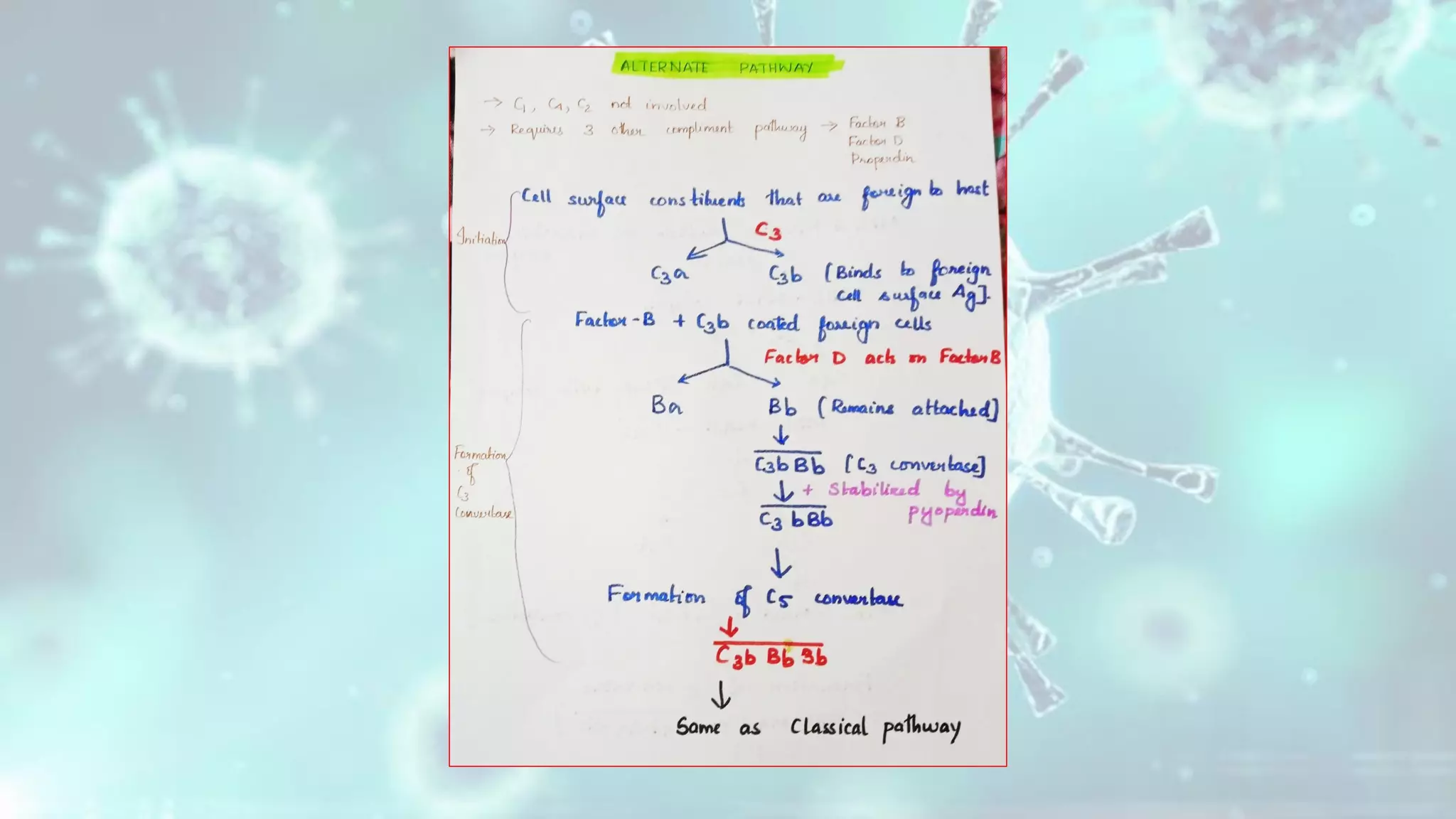
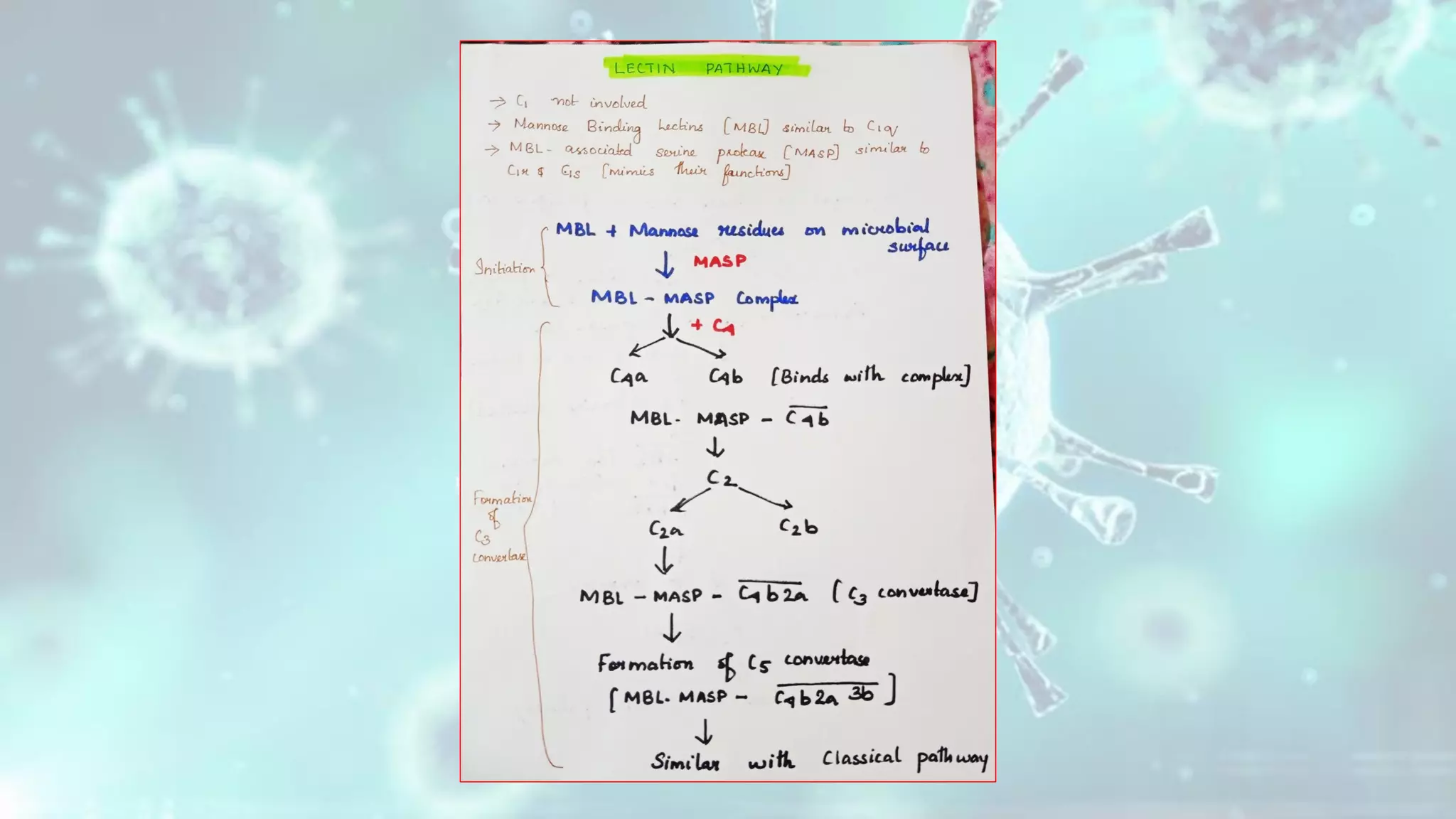
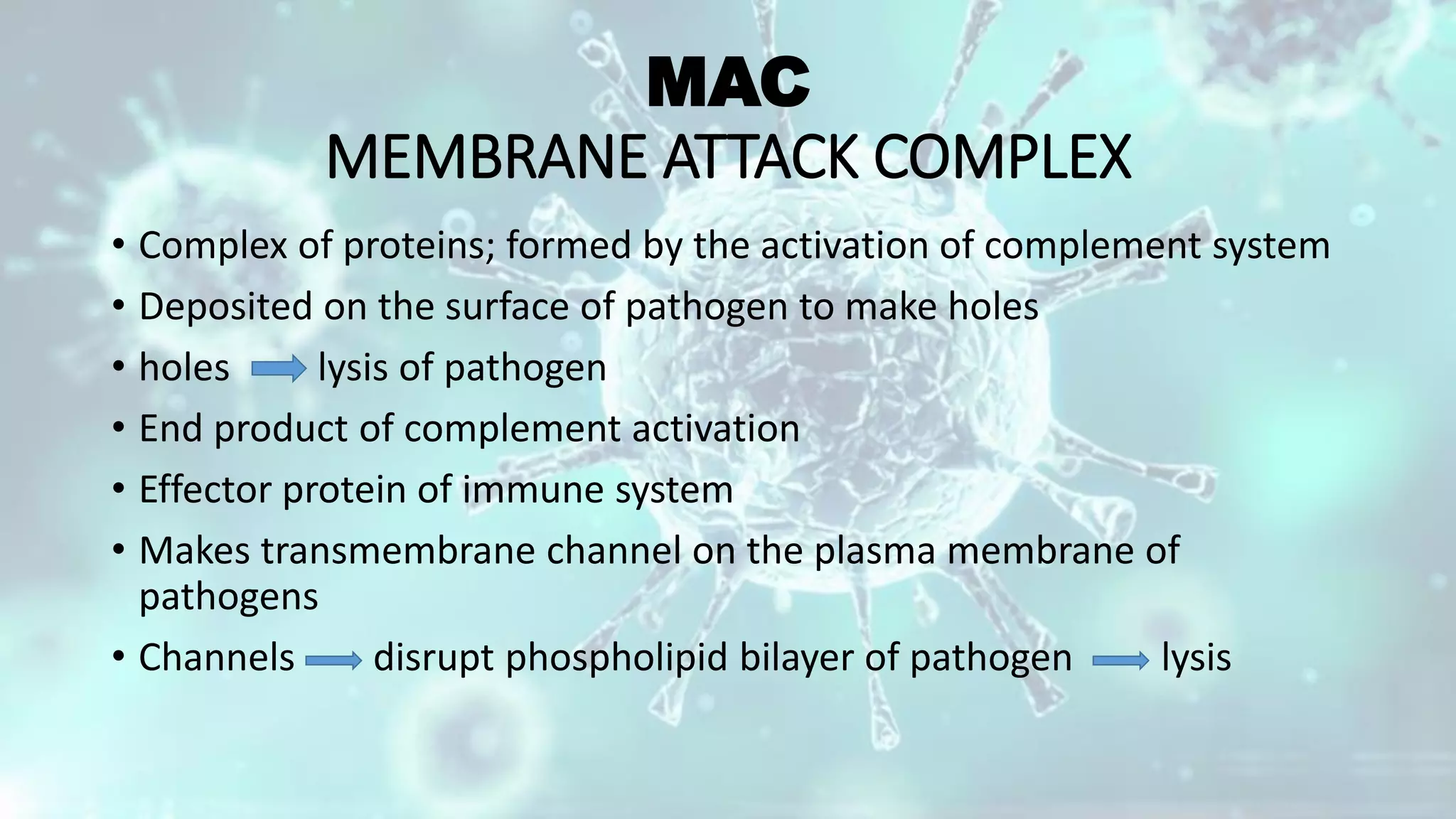
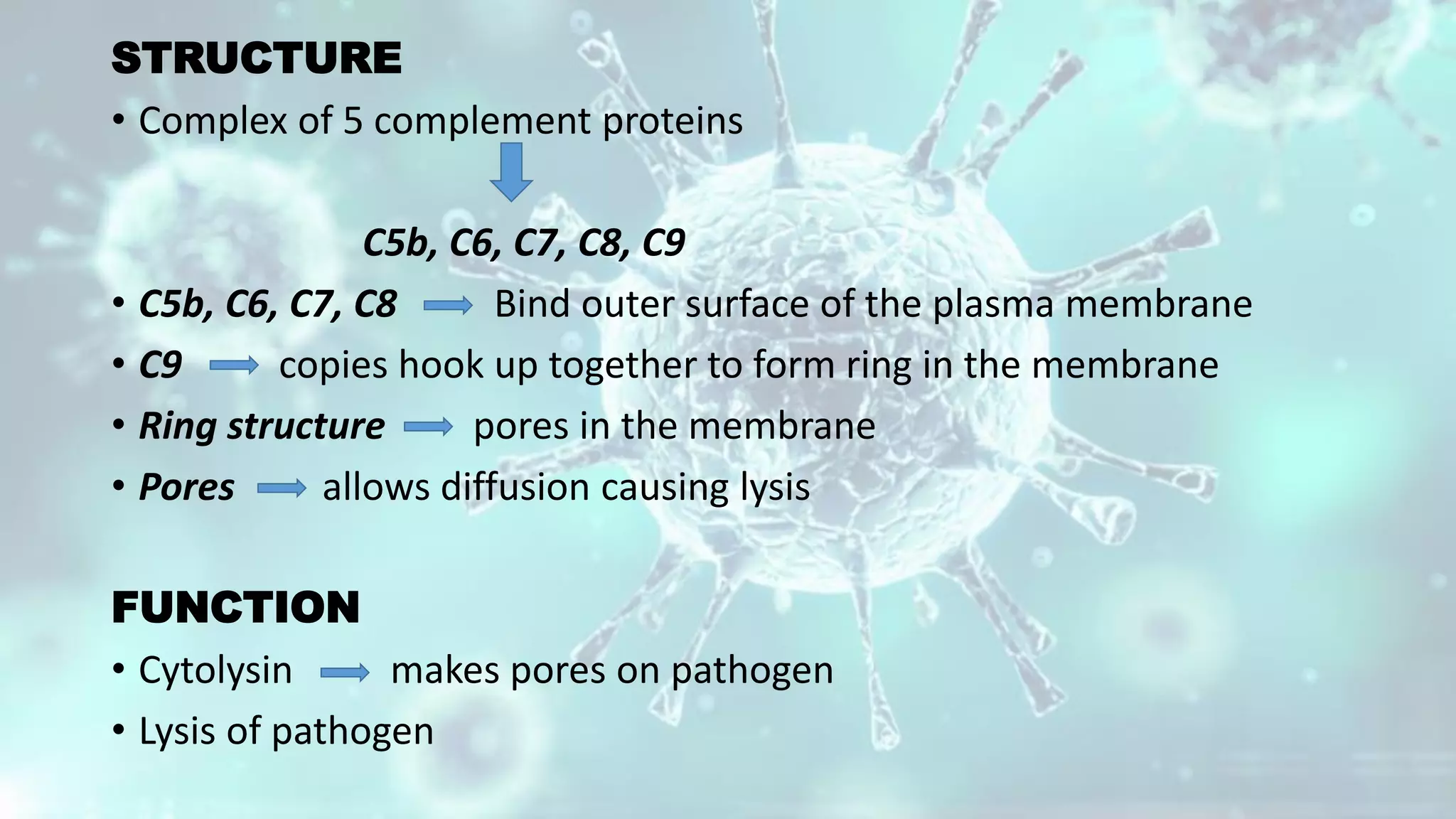
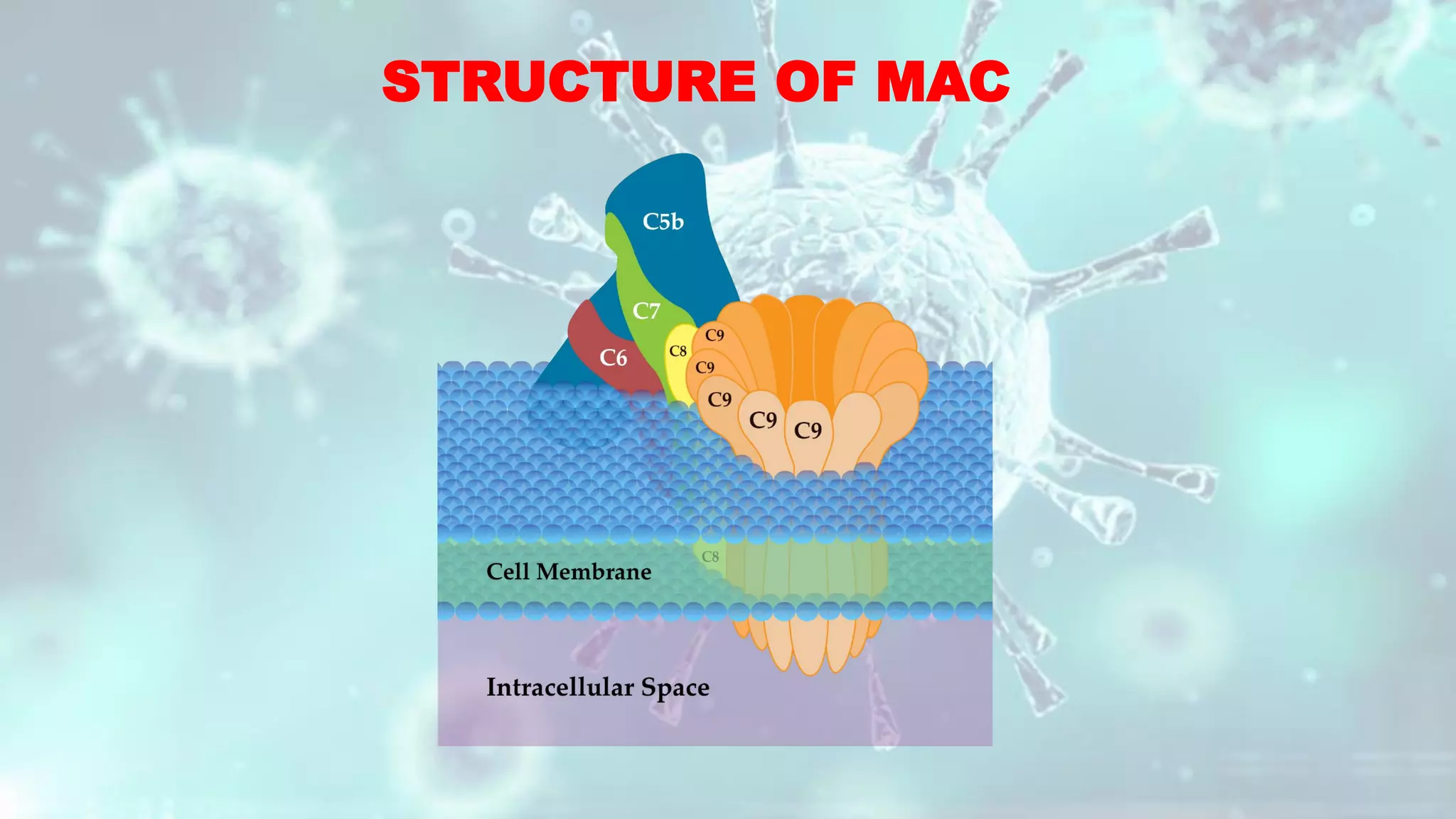
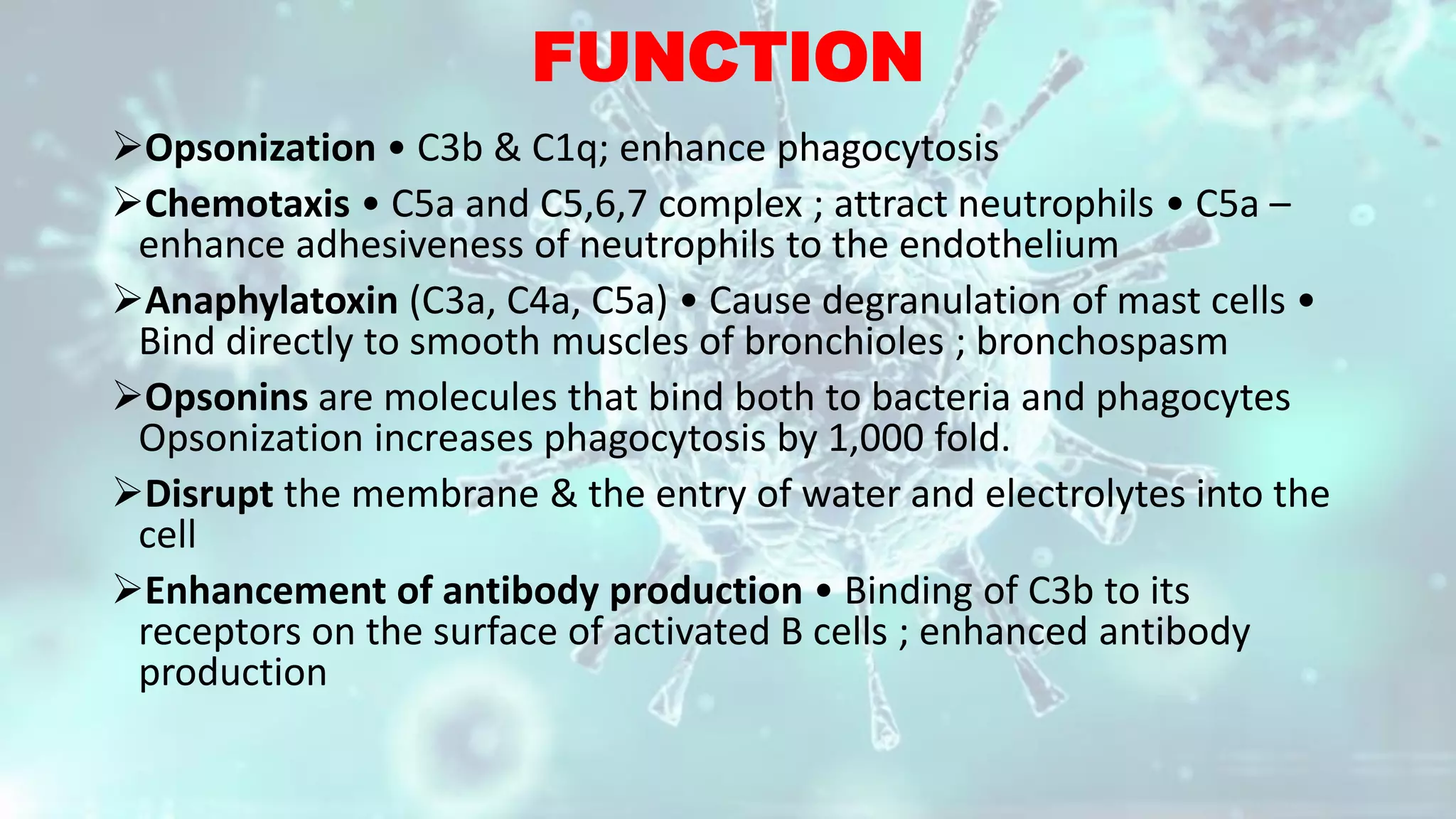
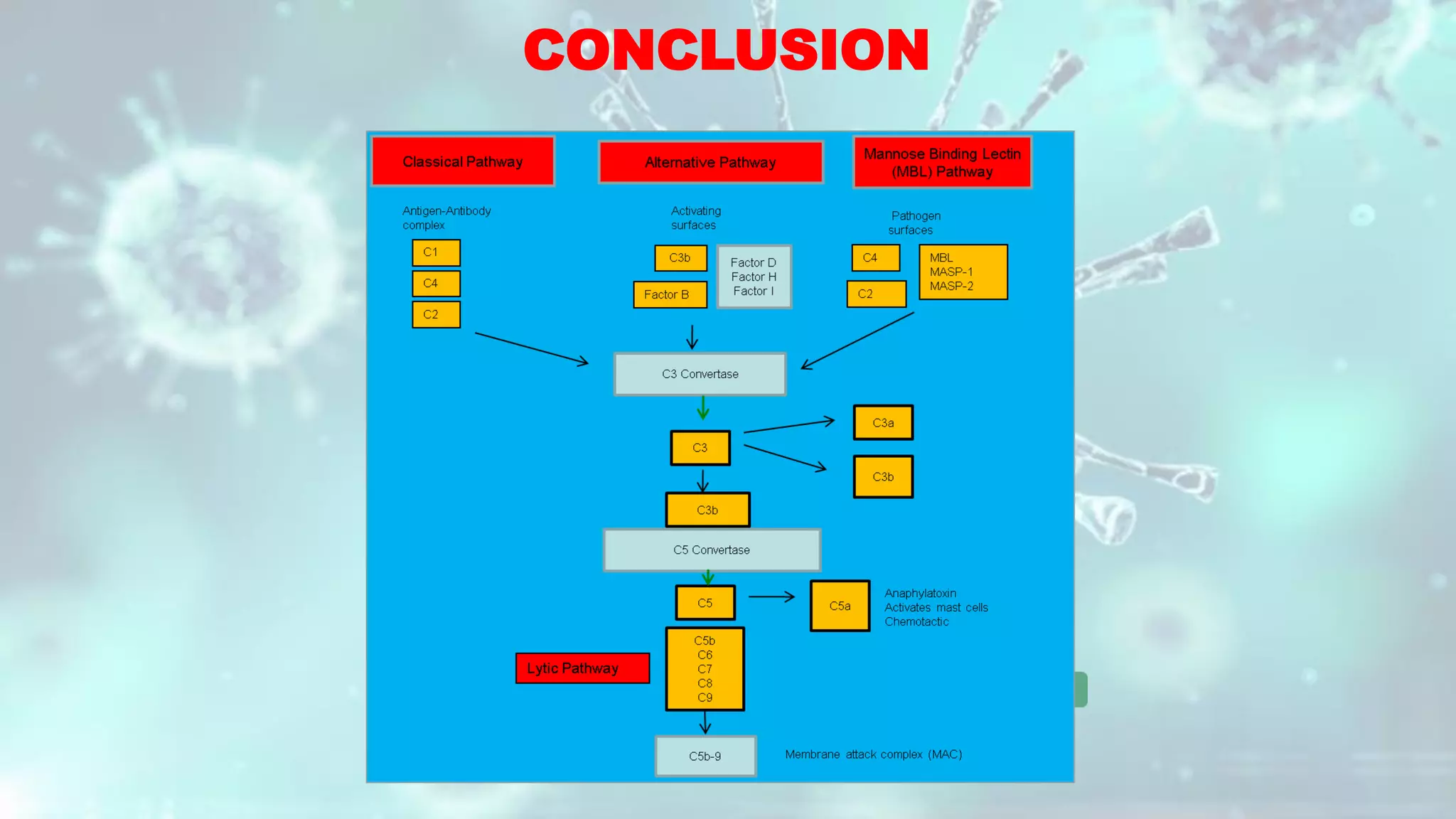
The document summarizes the complement system. It discusses the three complement pathways - classical, alternative, and lectin pathways. It also describes the structure and function of the membrane attack complex (MAC), which forms pores in pathogen cell membranes leading to lysis. The complement system plays an important role in innate immunity by opsonizing pathogens, attracting immune cells, and directly lysing certain pathogens.