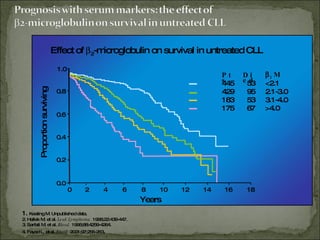
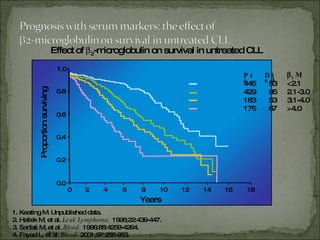
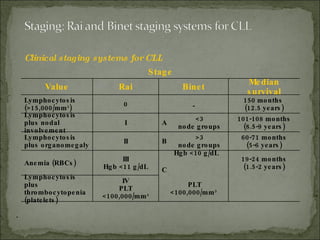
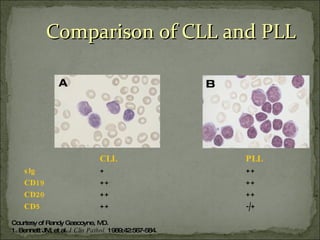
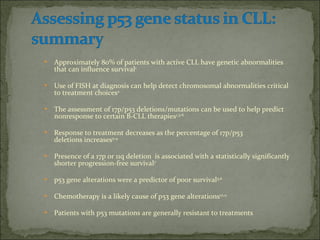
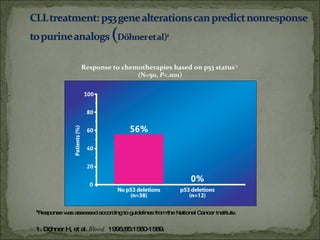
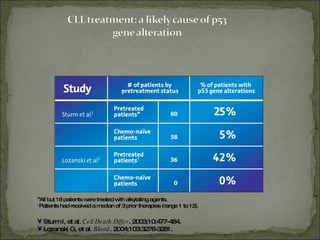
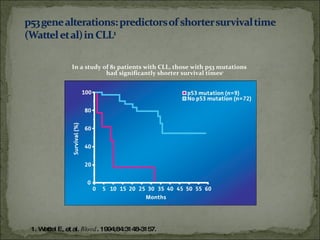
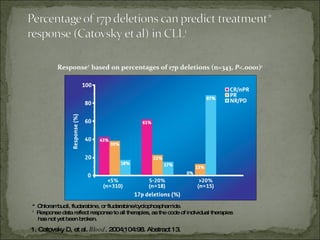
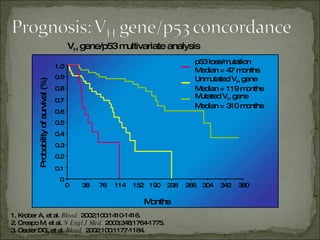
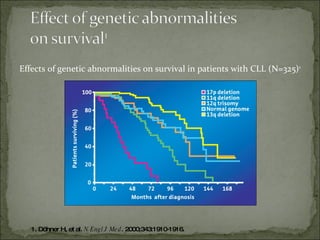
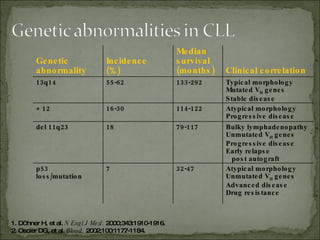
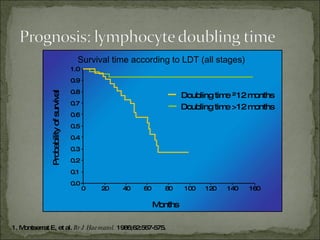
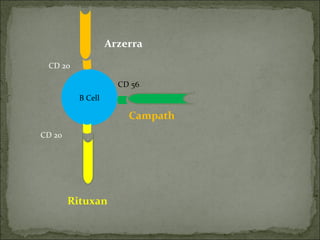
The document discusses the effects of β2-microglobulin and genetic markers on survival in untreated chronic lymphocytic leukemia (CLL) patients, highlighting the significant impact of genetic abnormalities such as p53 mutations on treatment response and progression-free survival. It outlines clinical staging systems, compares CLL with prolymphocytic leukemia (PLL), and emphasizes the importance of proper diagnosis and treatment strategies. Genetic markers are increasingly recognized for their predictive value in disease course and survival outcomes.