This document summarizes several key organelles and structures within cells:
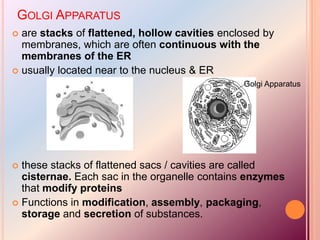
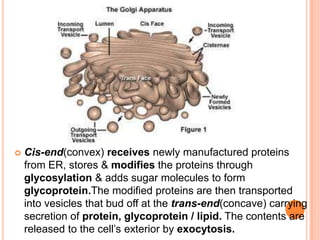
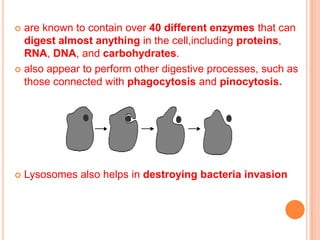
- The Golgi apparatus stacks flattened sacs that modify and package proteins and lipids for secretion or transport within the cell. Lysosomes contain digestive enzymes and help break down materials through autophagy and phagocytosis.
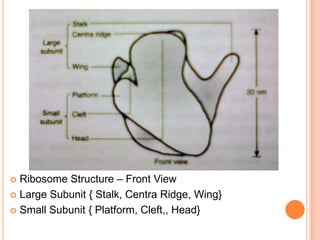
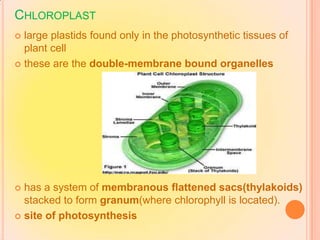
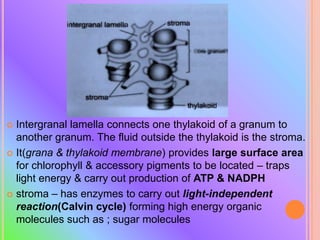
- Ribosomes are composed of RNA and proteins and are the site of protein synthesis. Chloroplasts contain chlorophyll and carry out photosynthesis through their thylakoid membranes and grana stacks in plant cells.
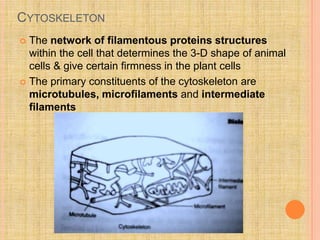
- The cytoskeleton consists of microtubules, microfilaments, and intermediate filaments that give cells their shape and allow for cellular movement and transport.