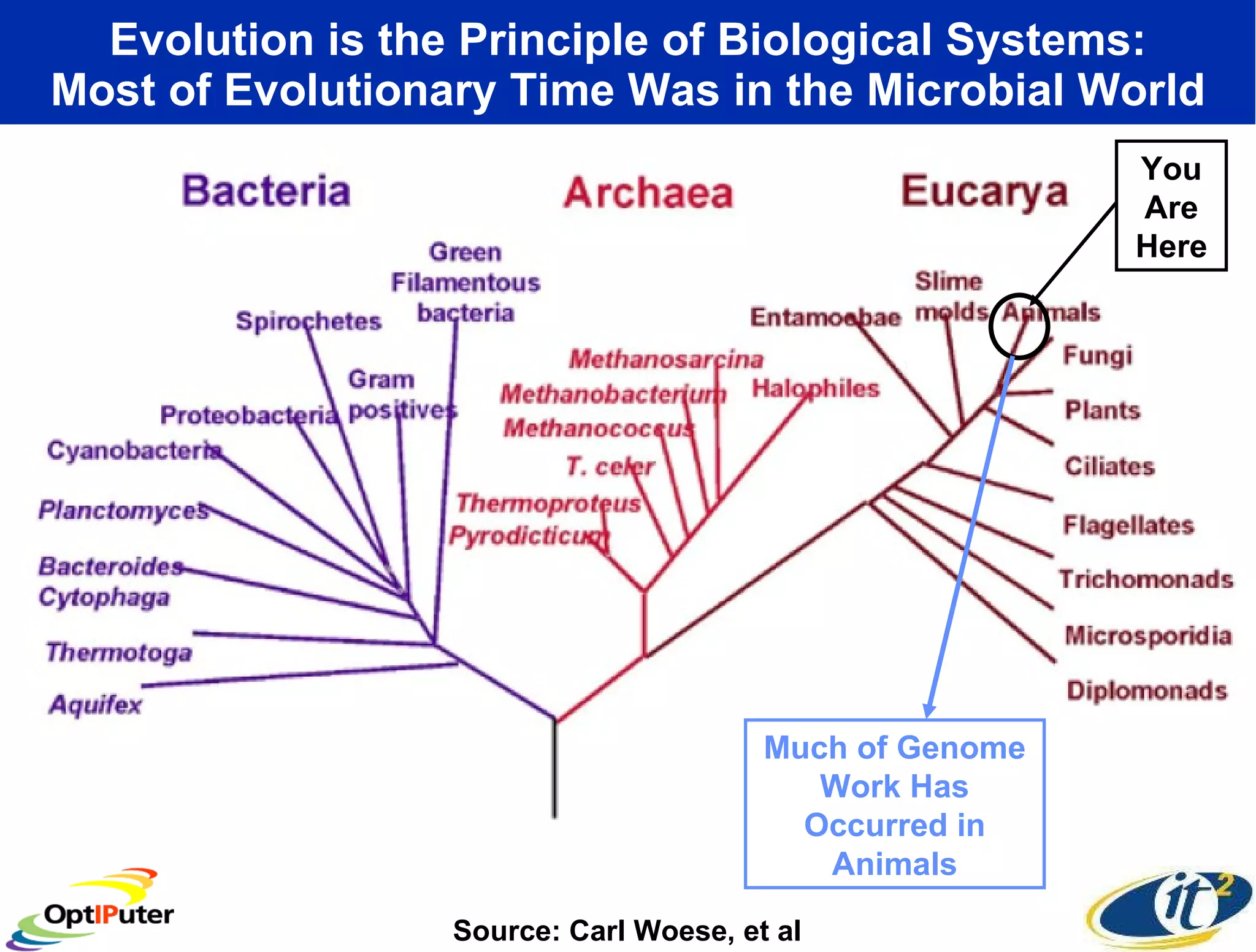
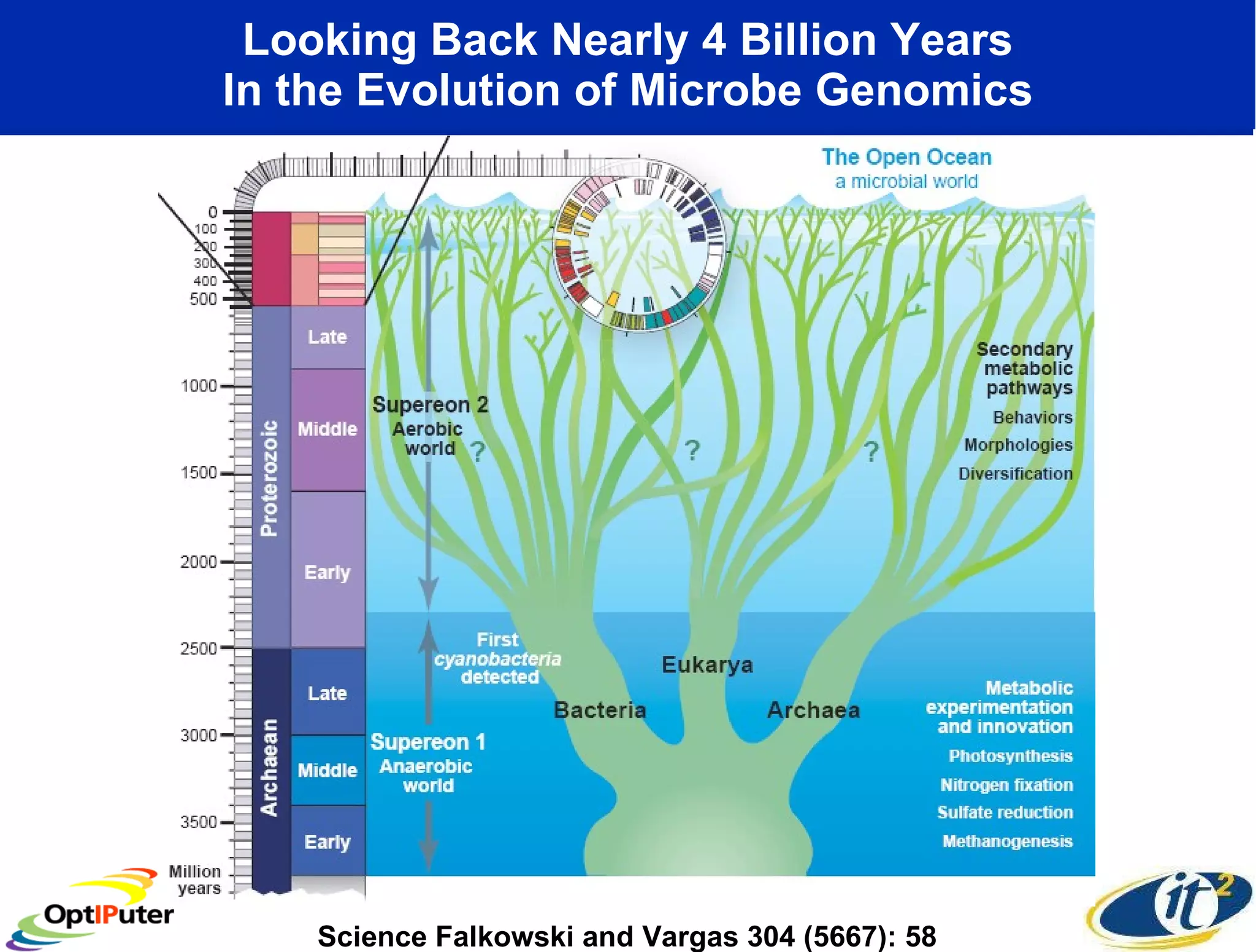
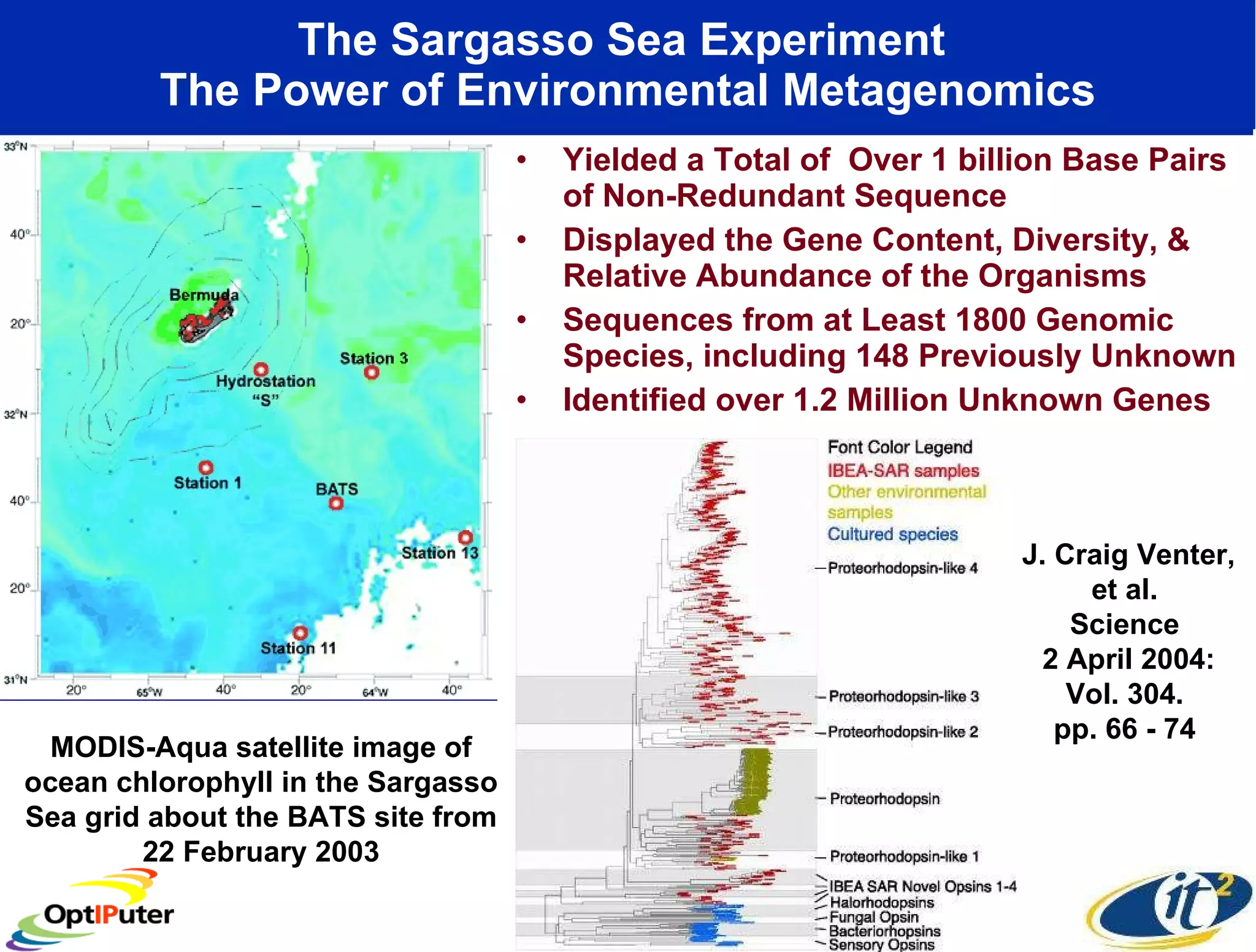
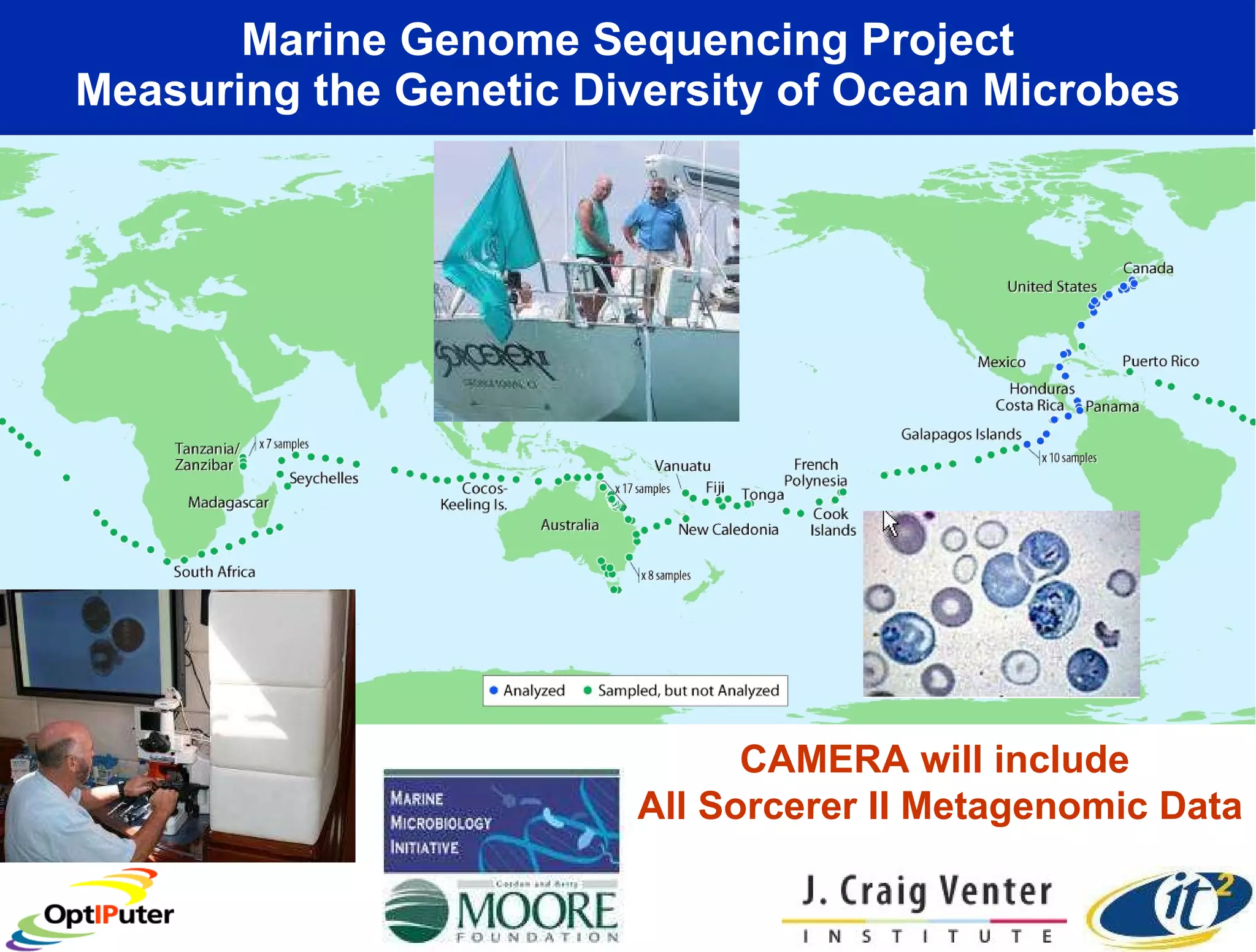
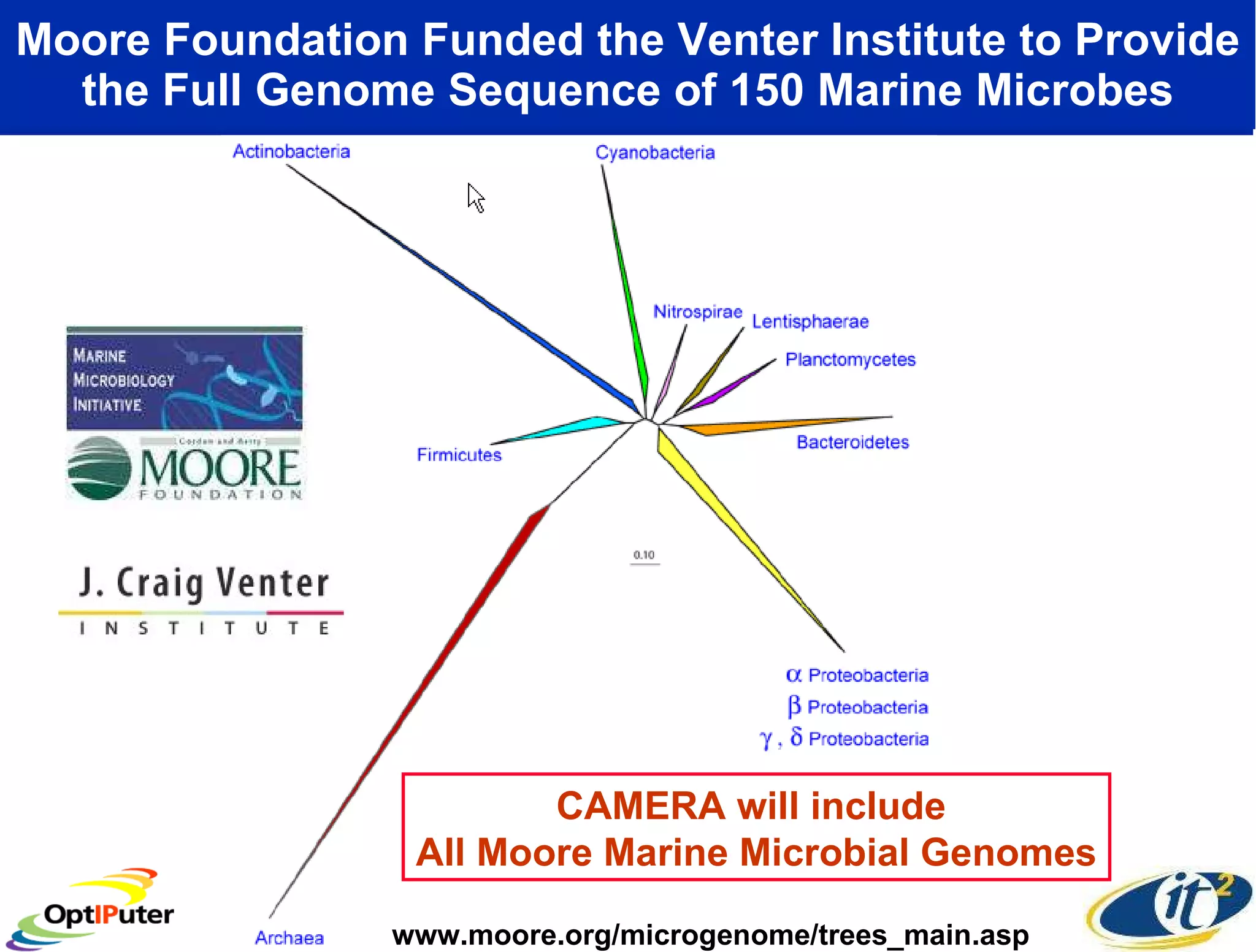
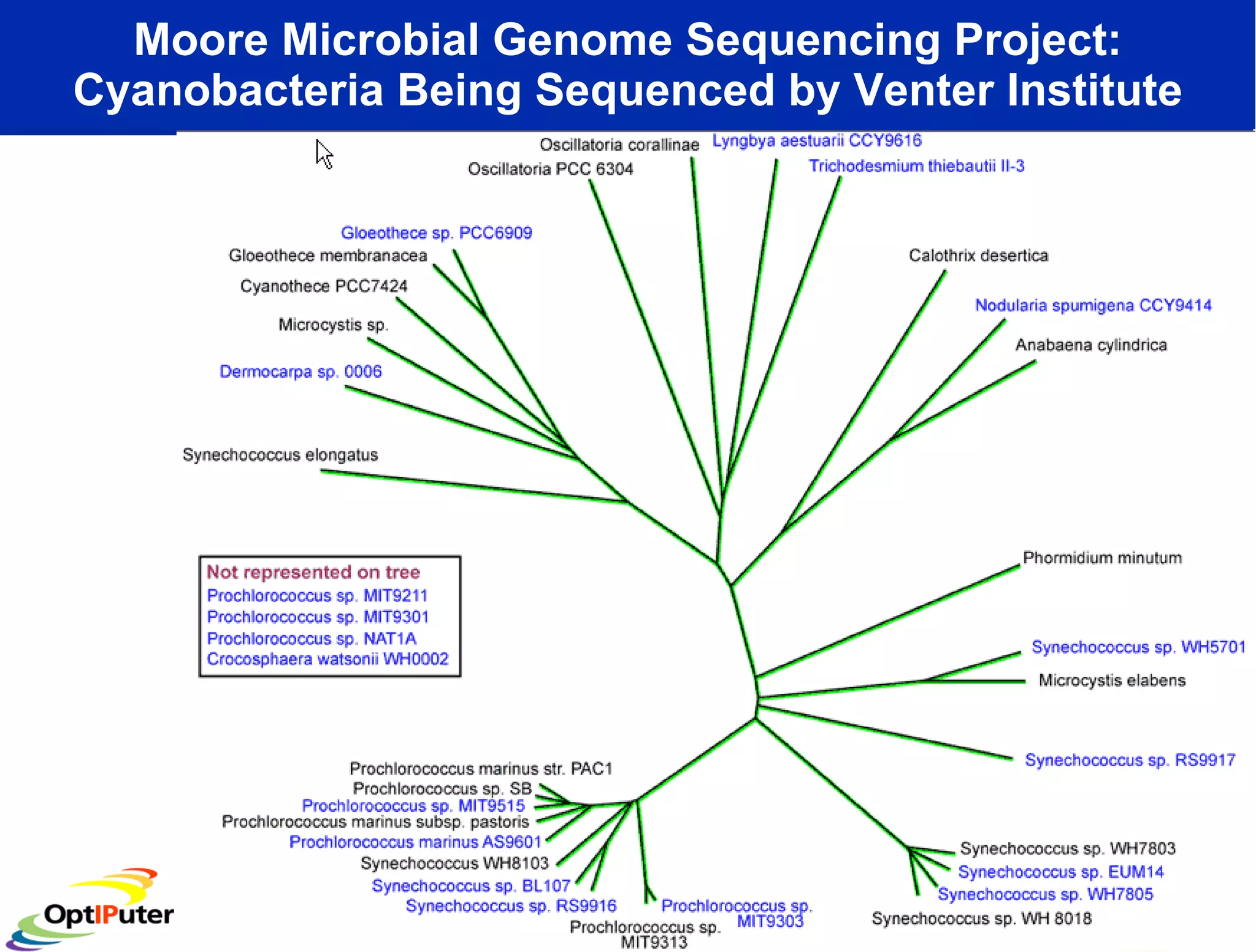
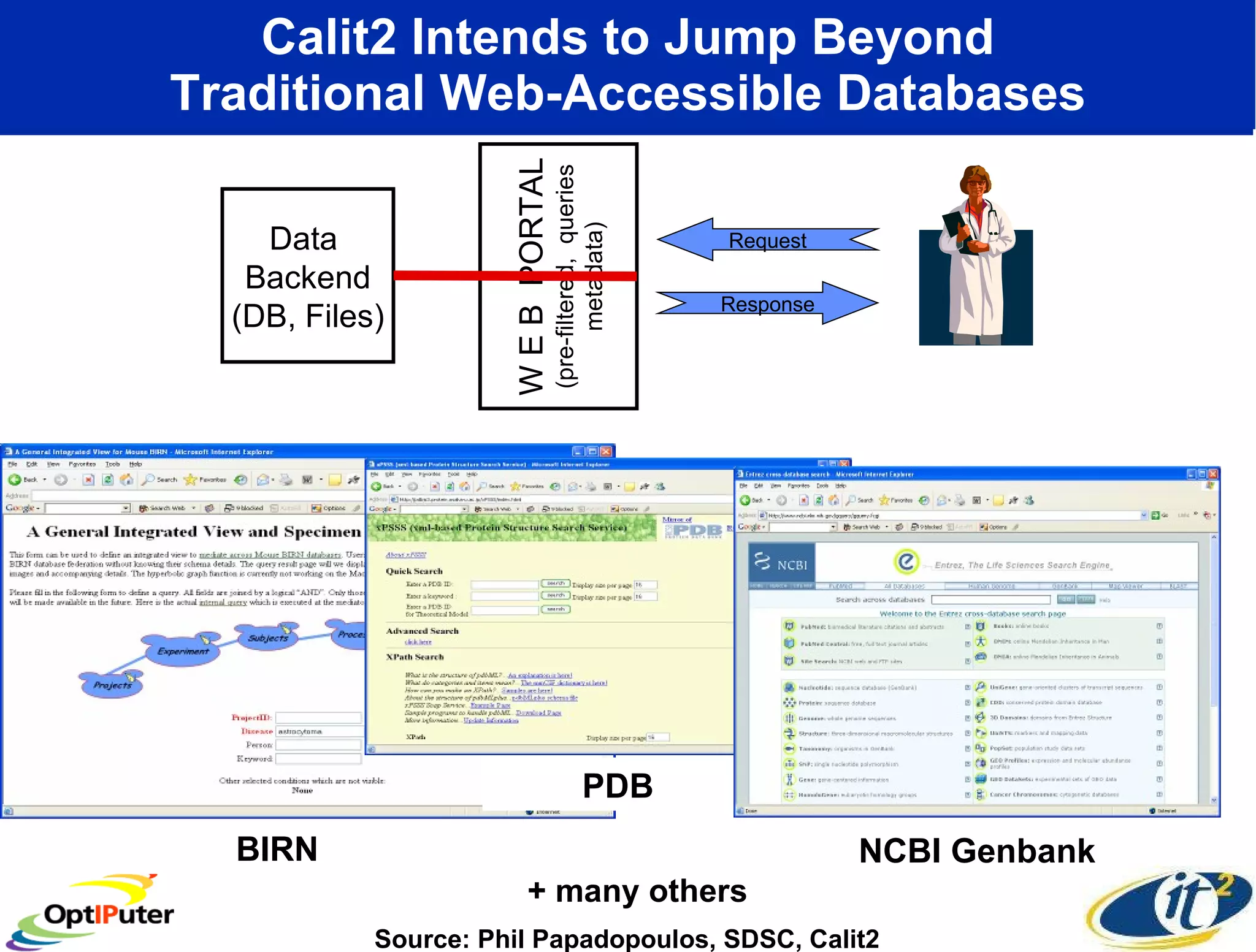
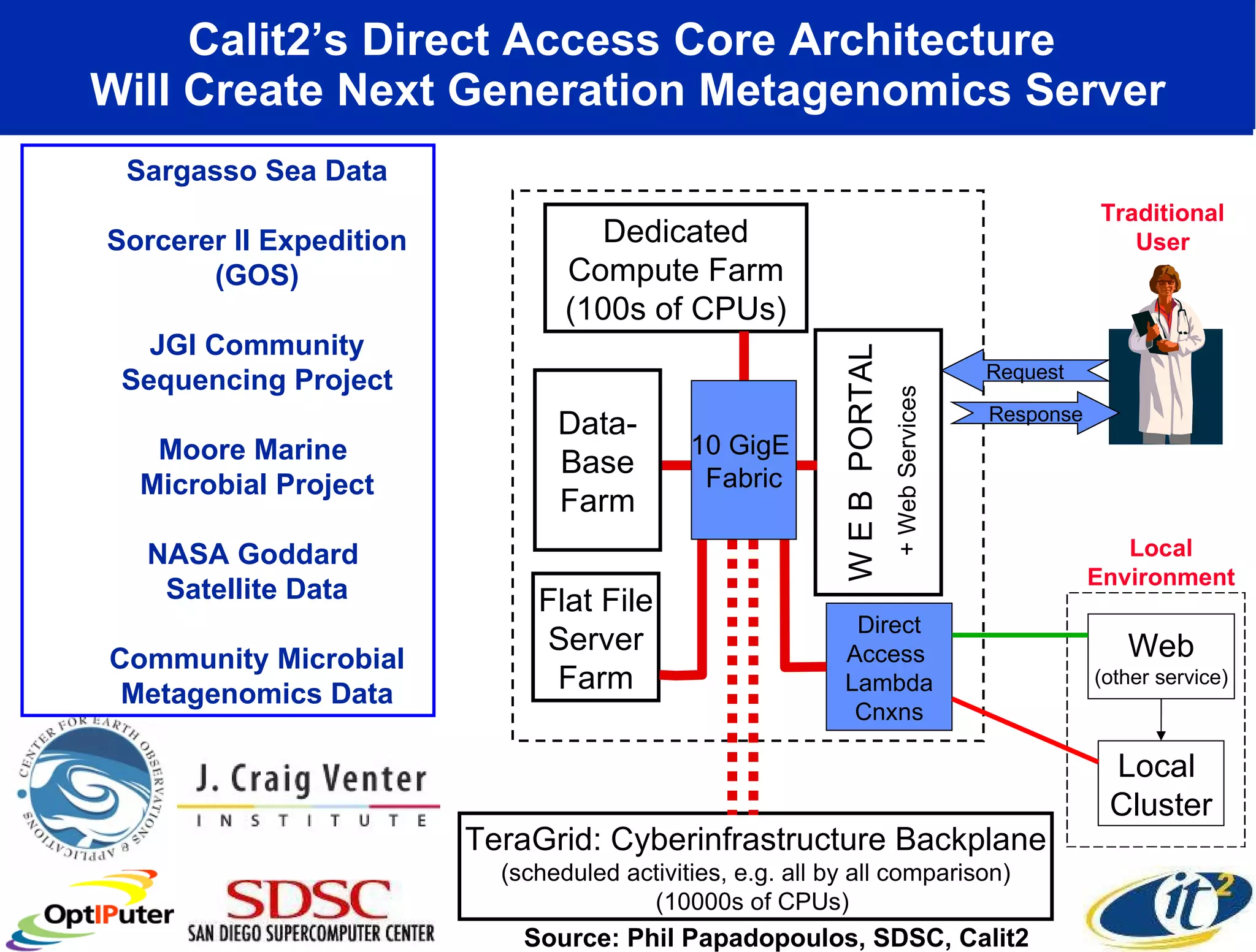
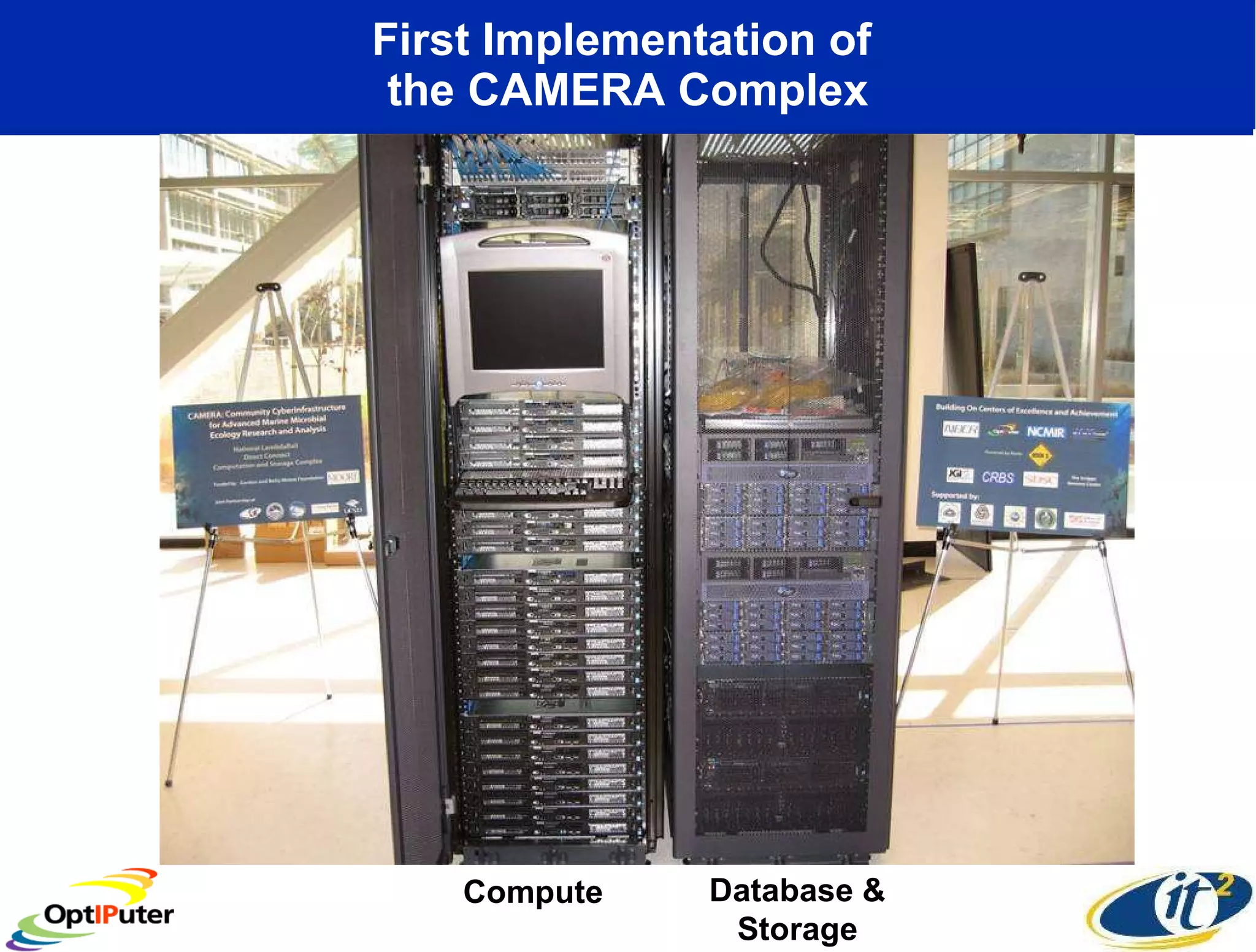
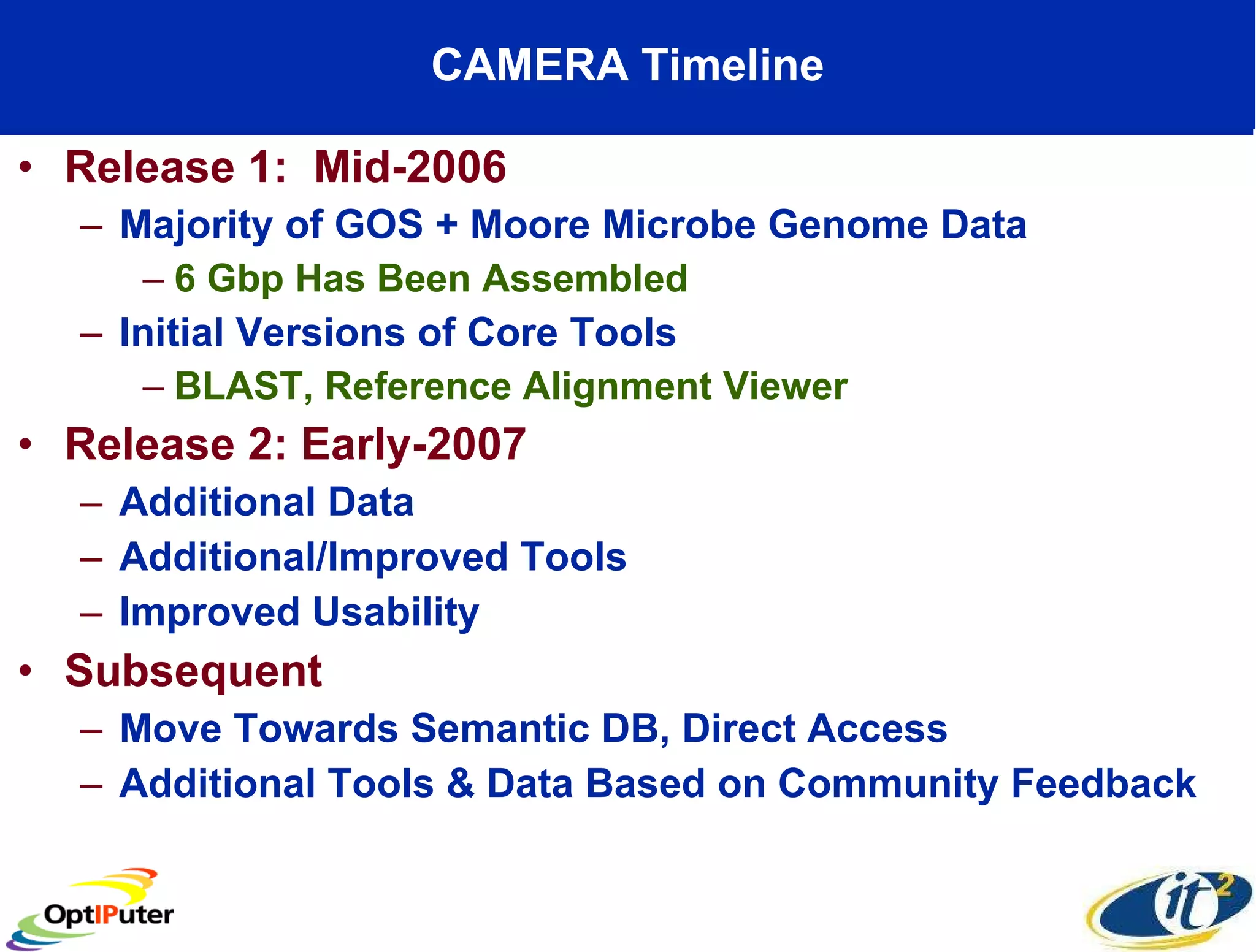
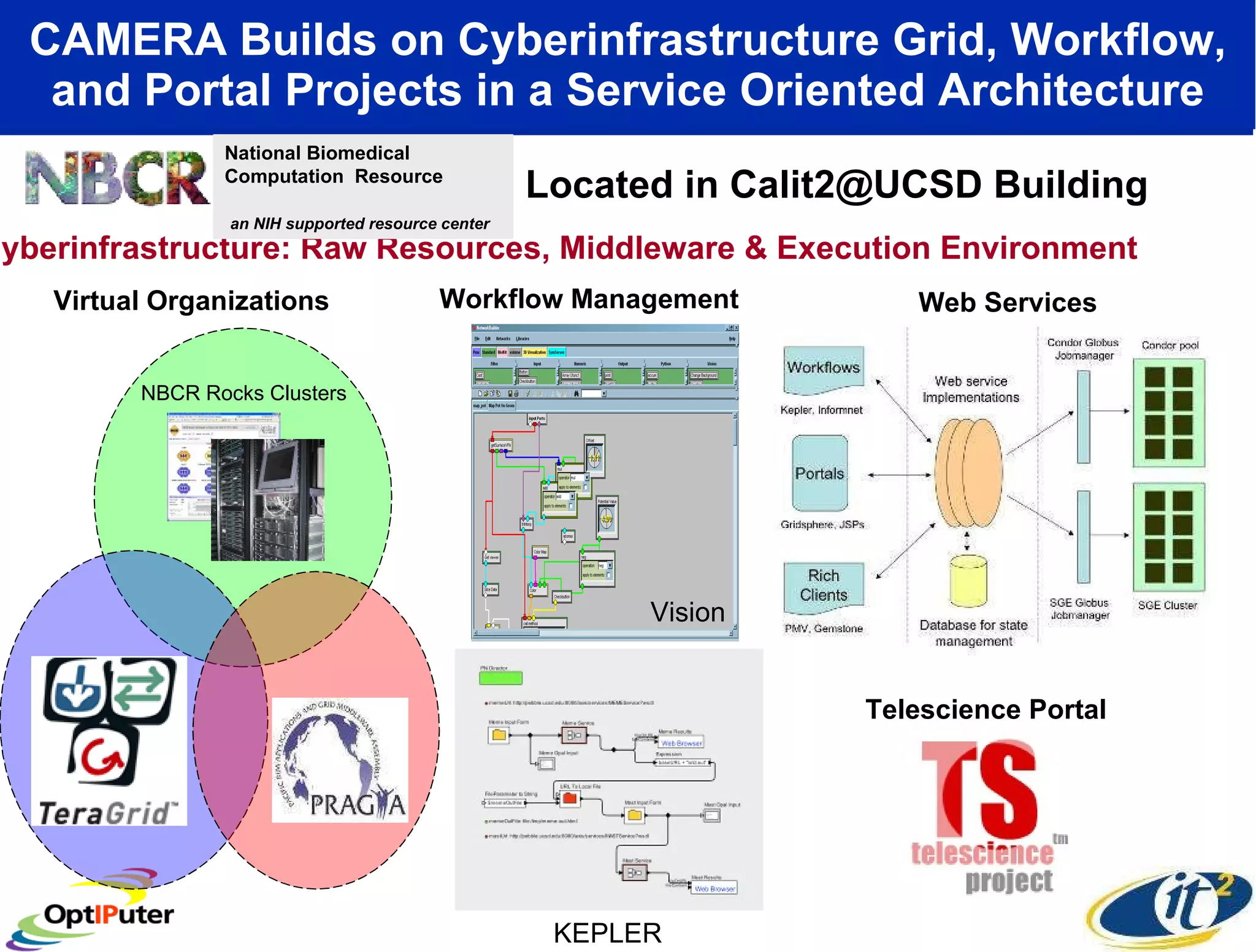
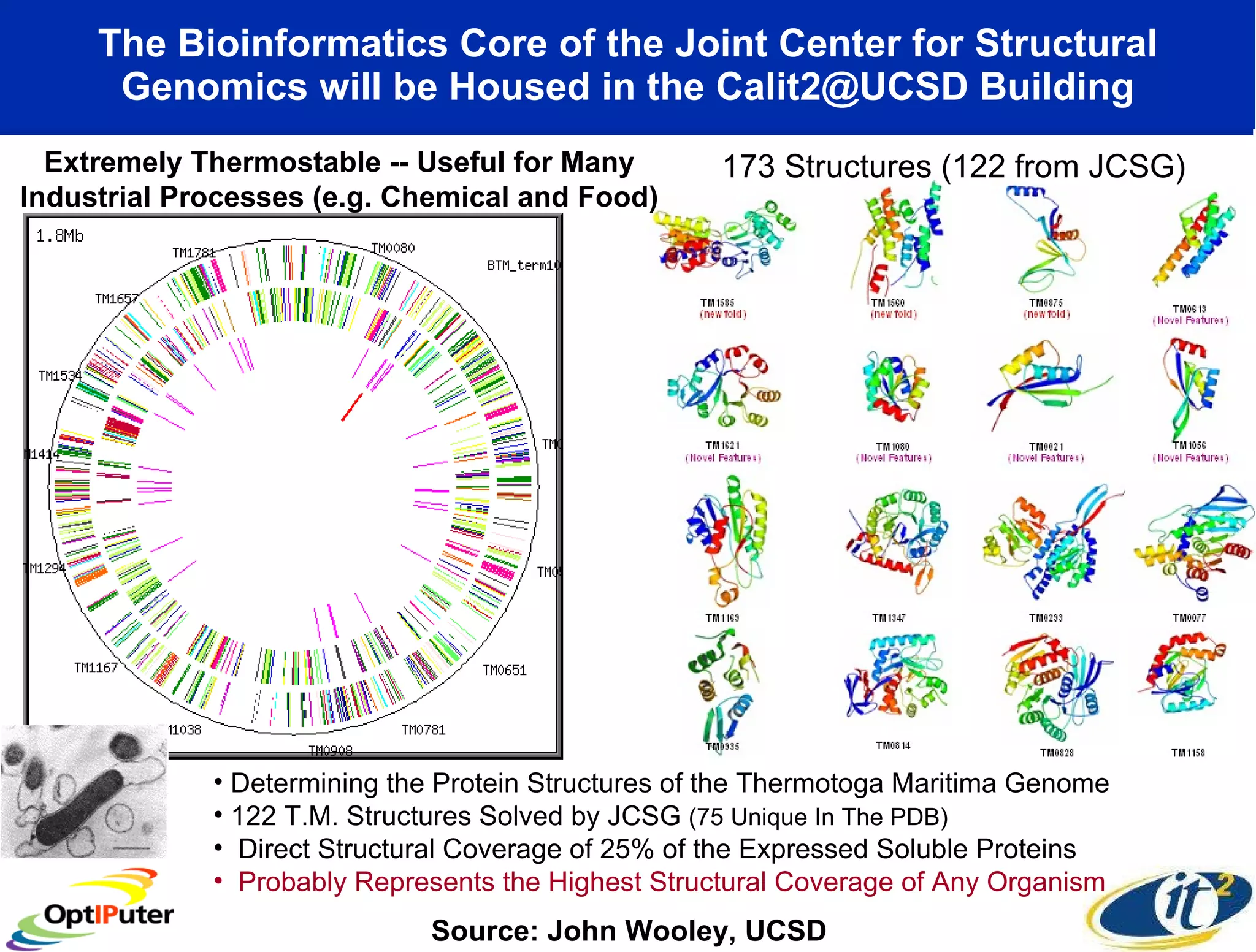
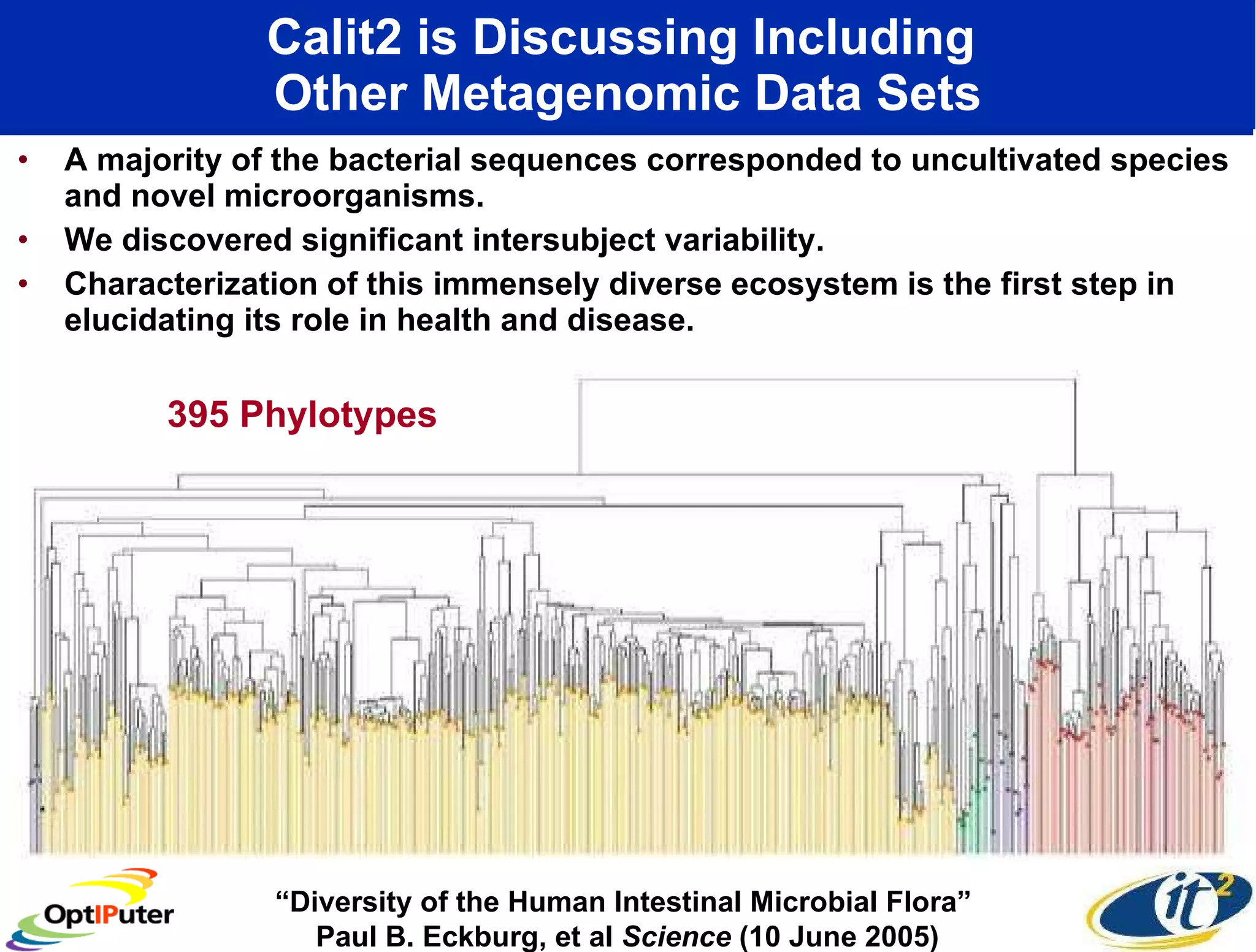
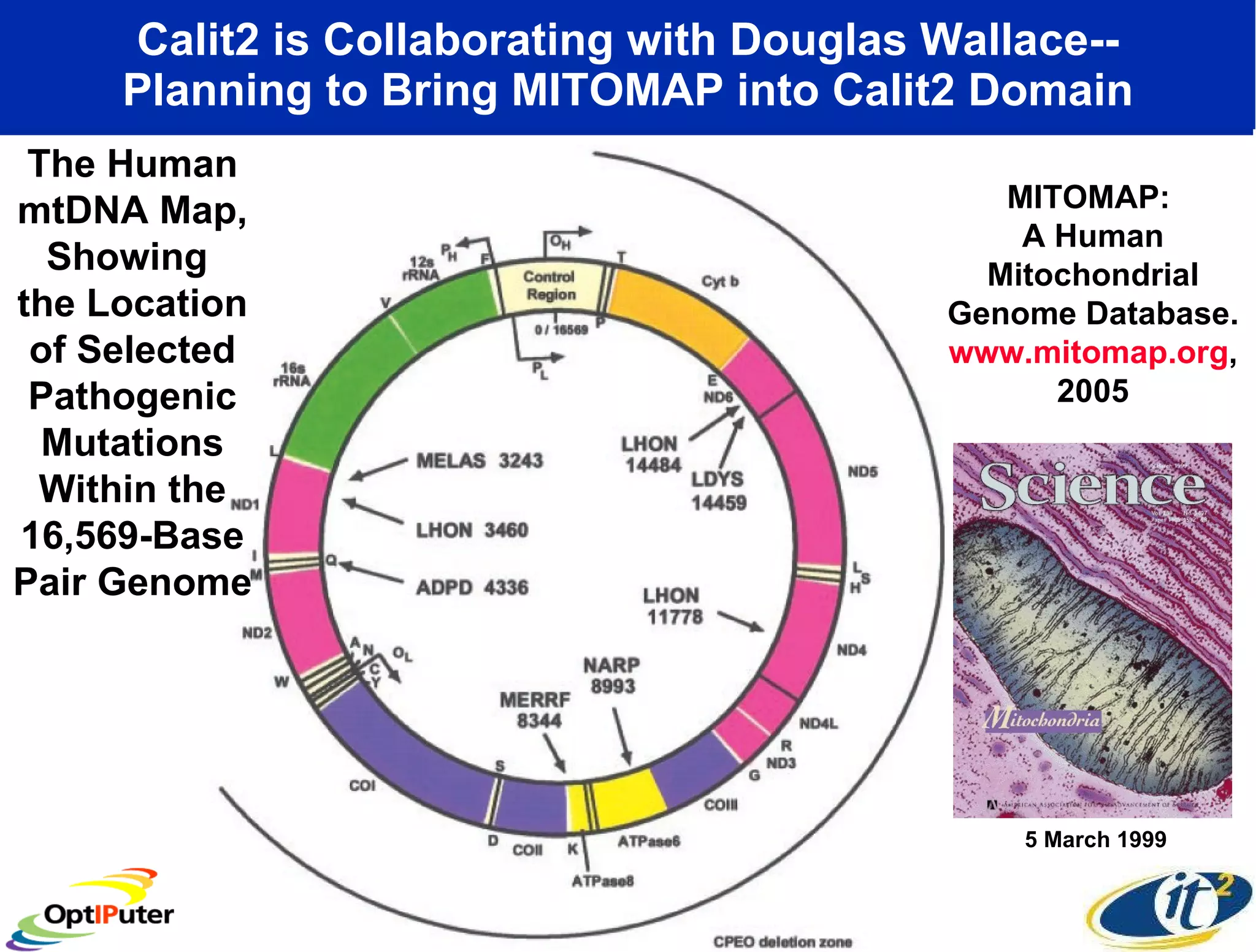
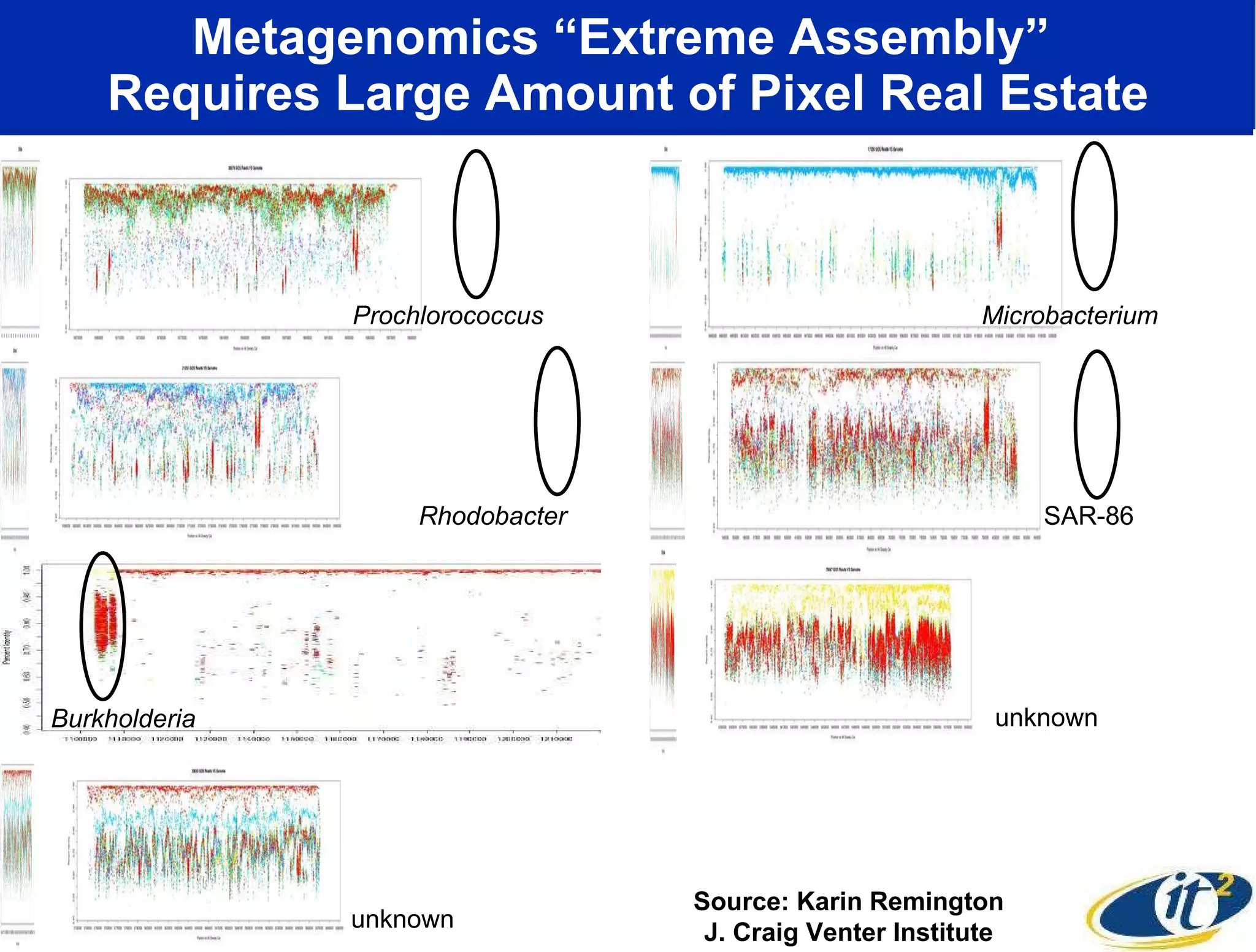
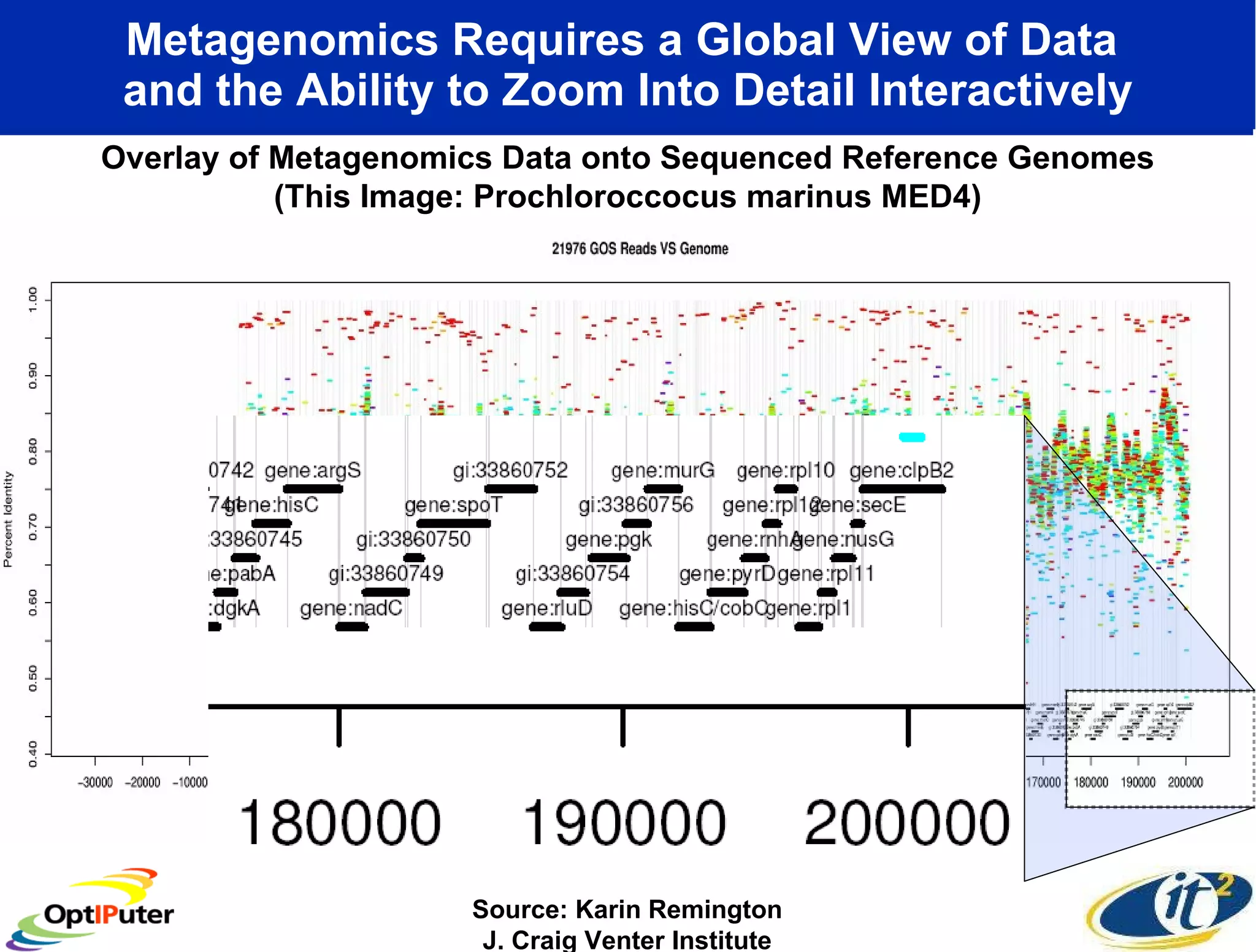
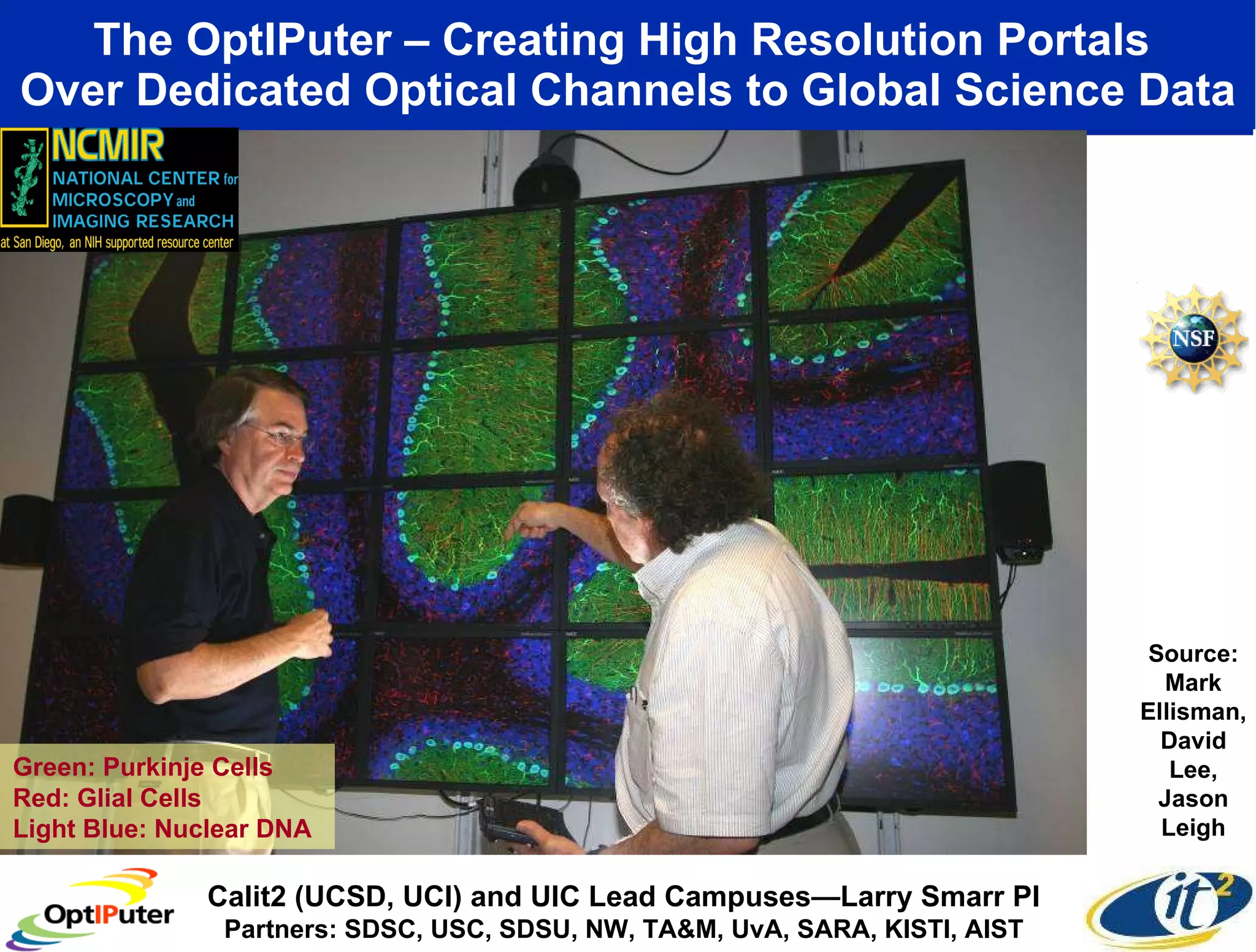
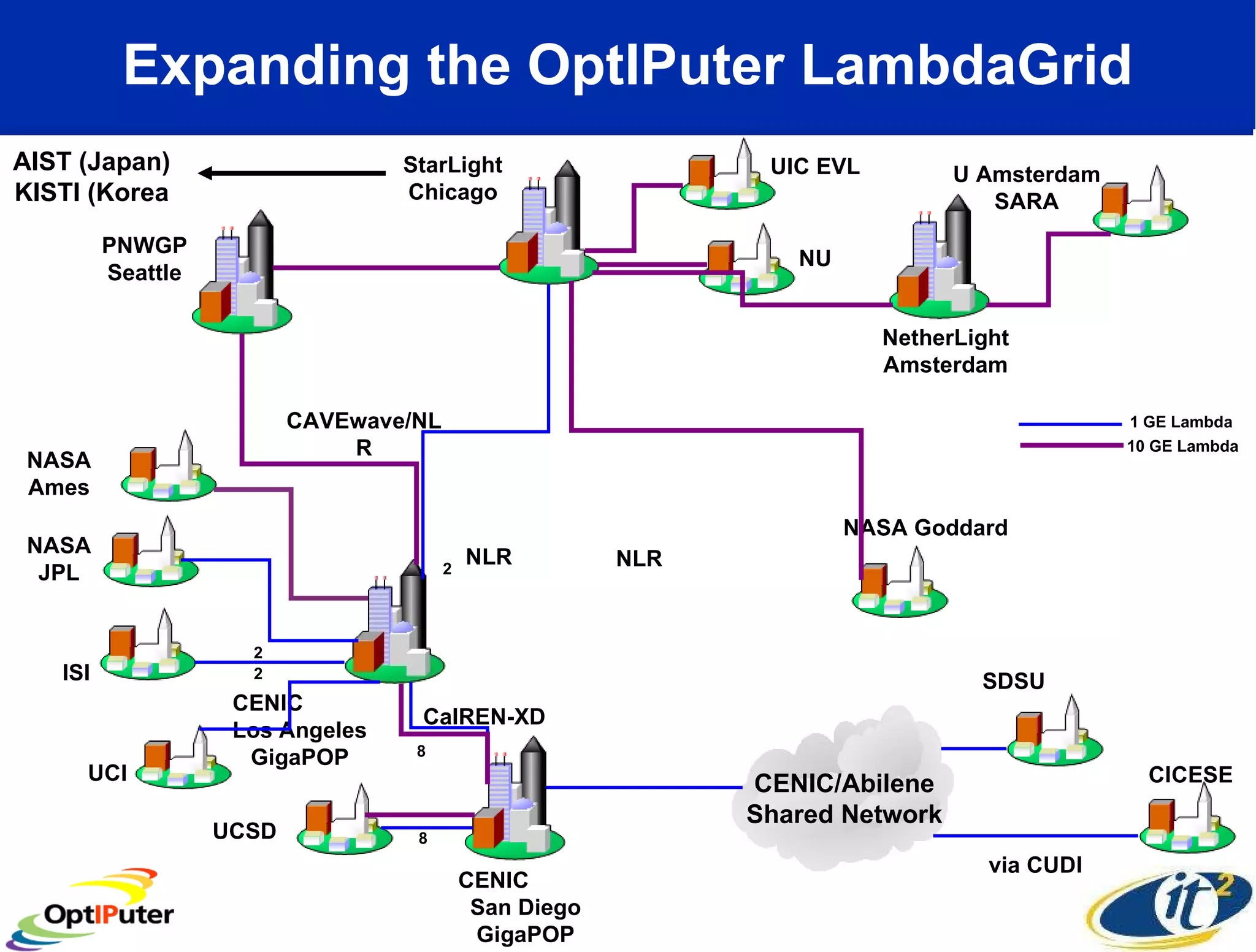
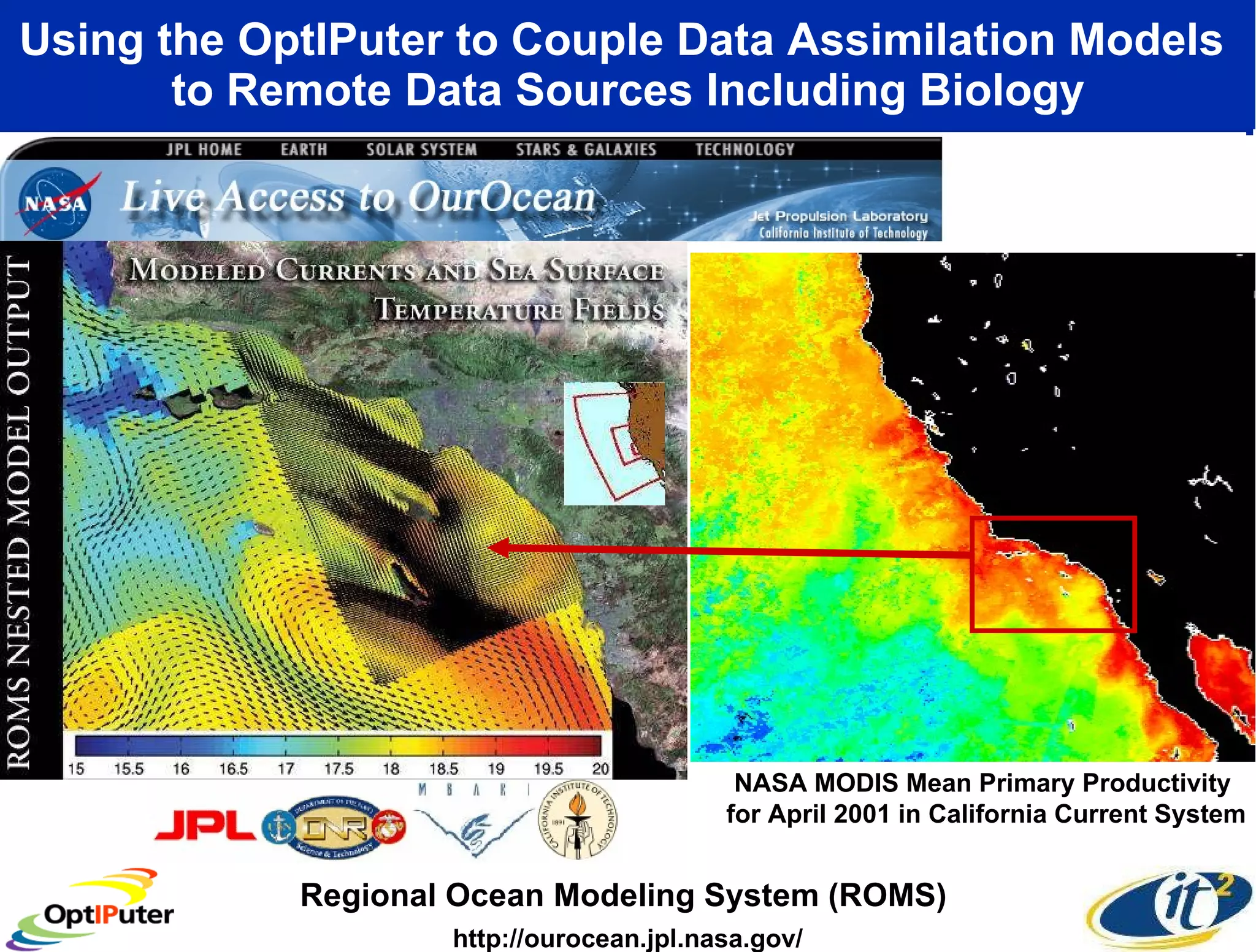
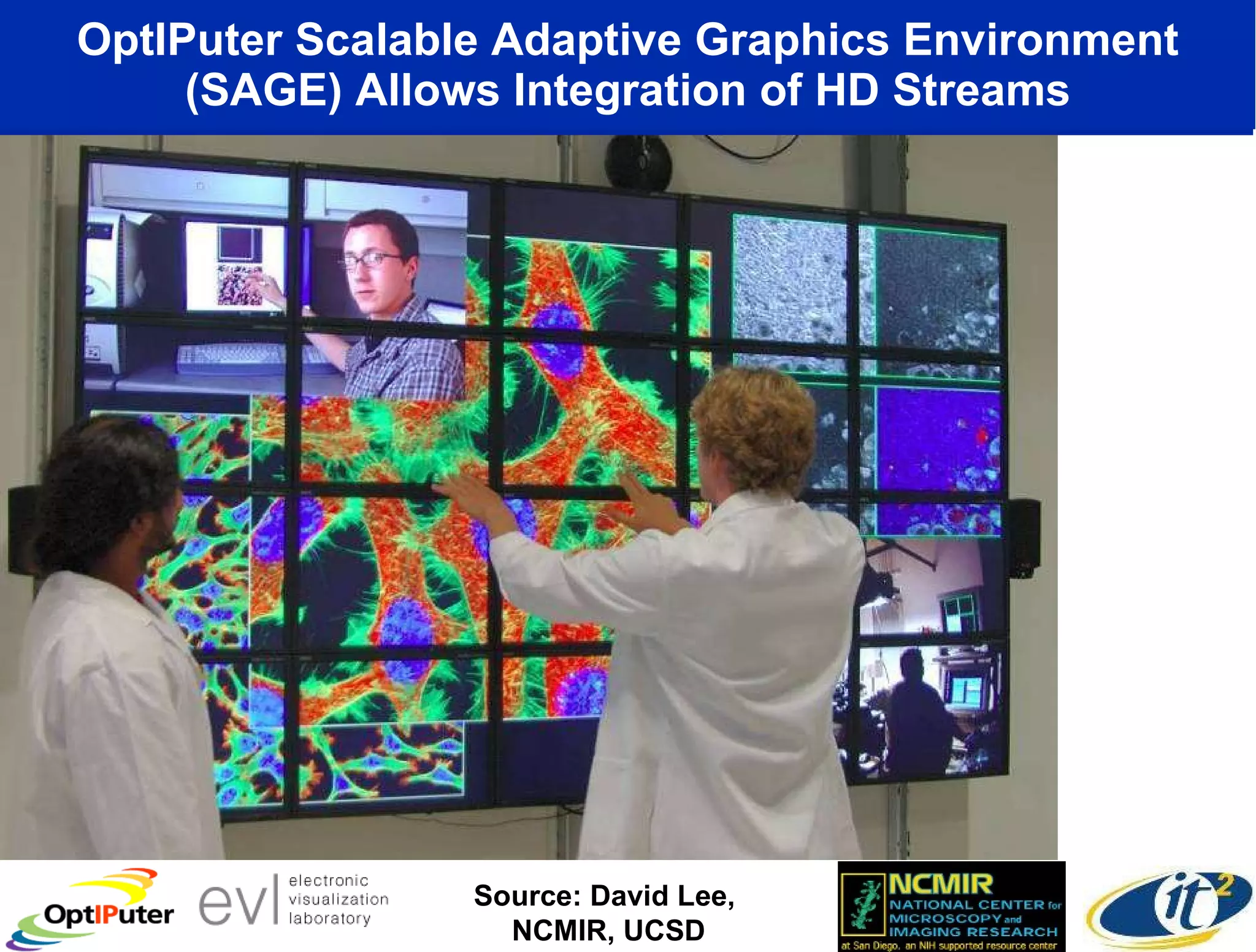
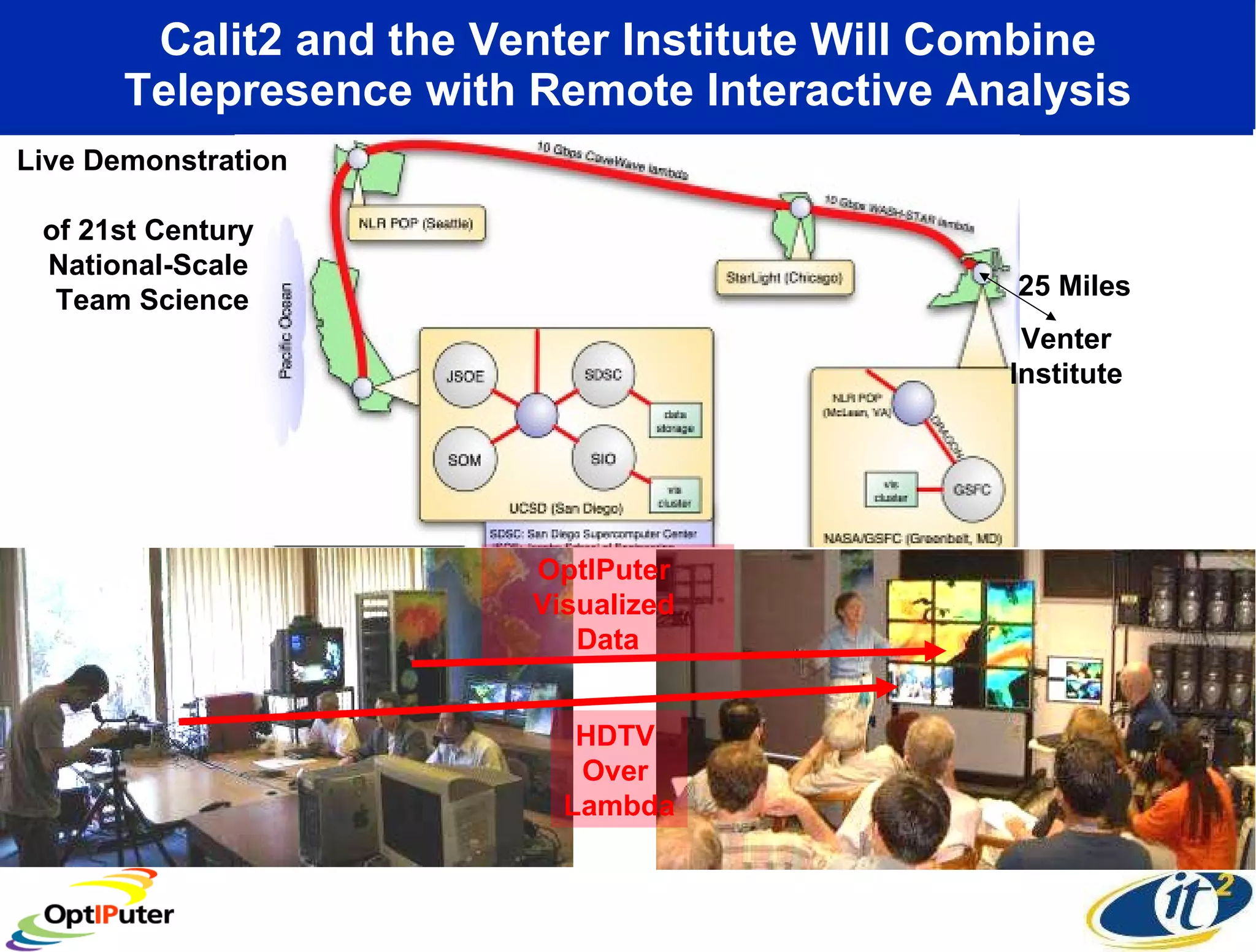
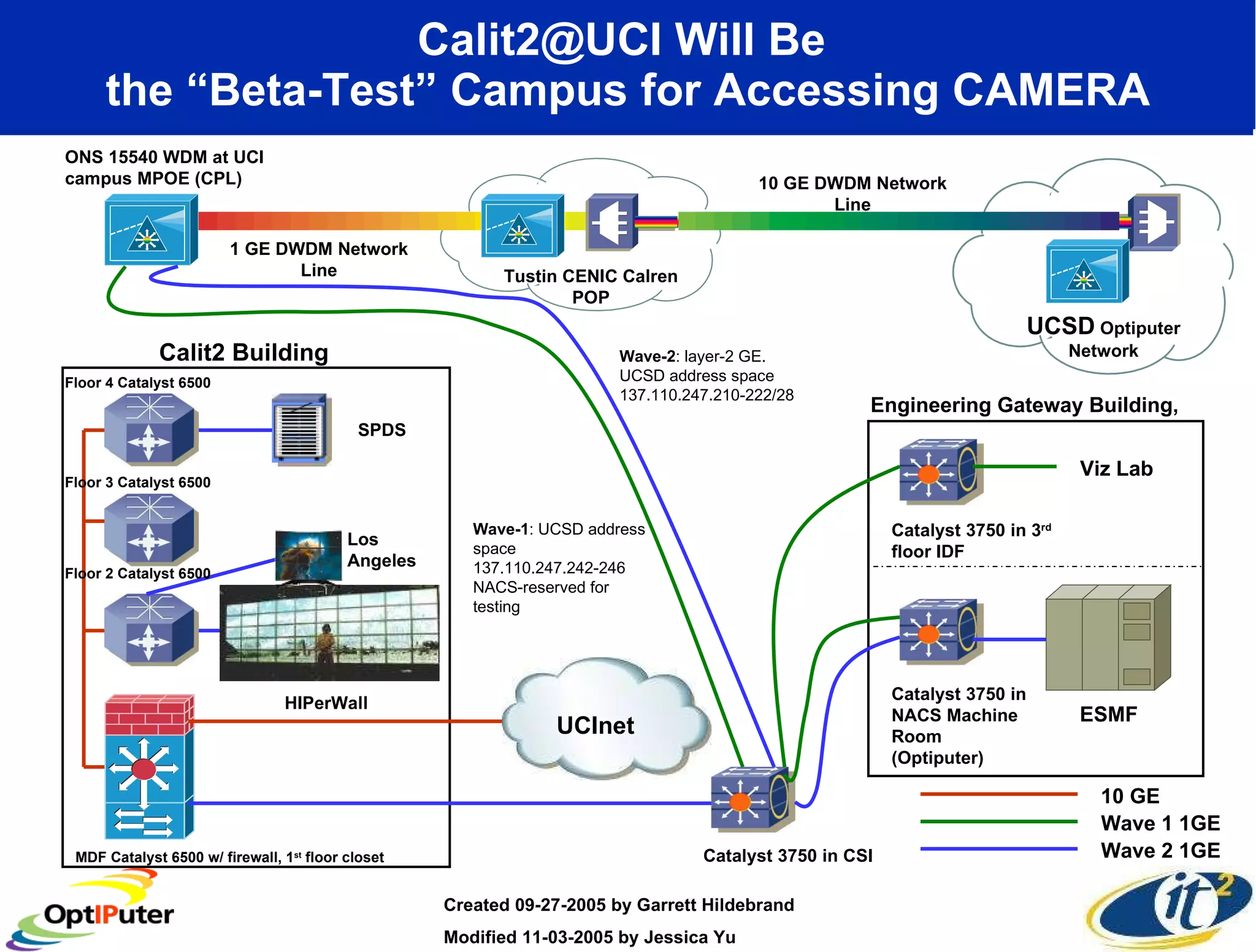
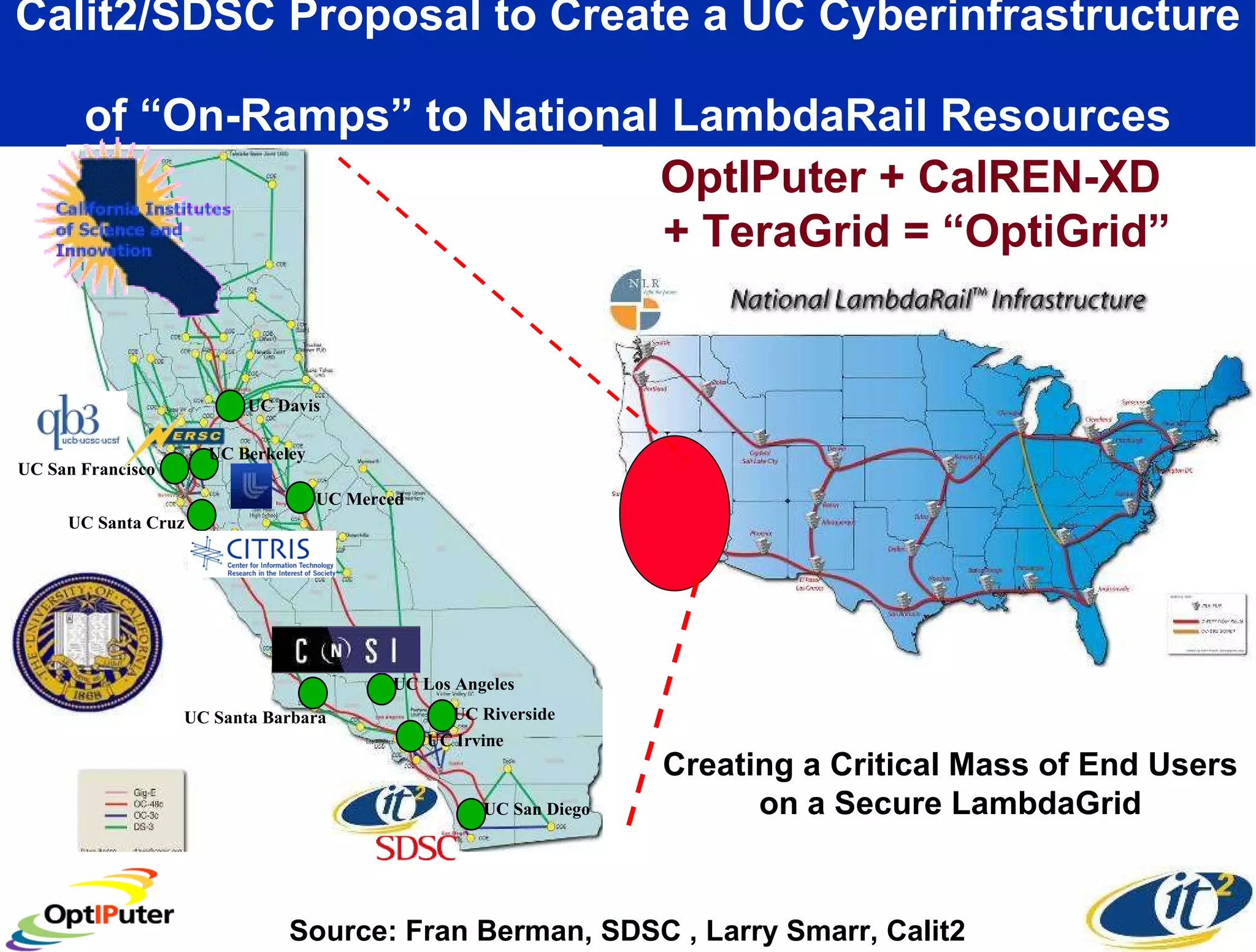
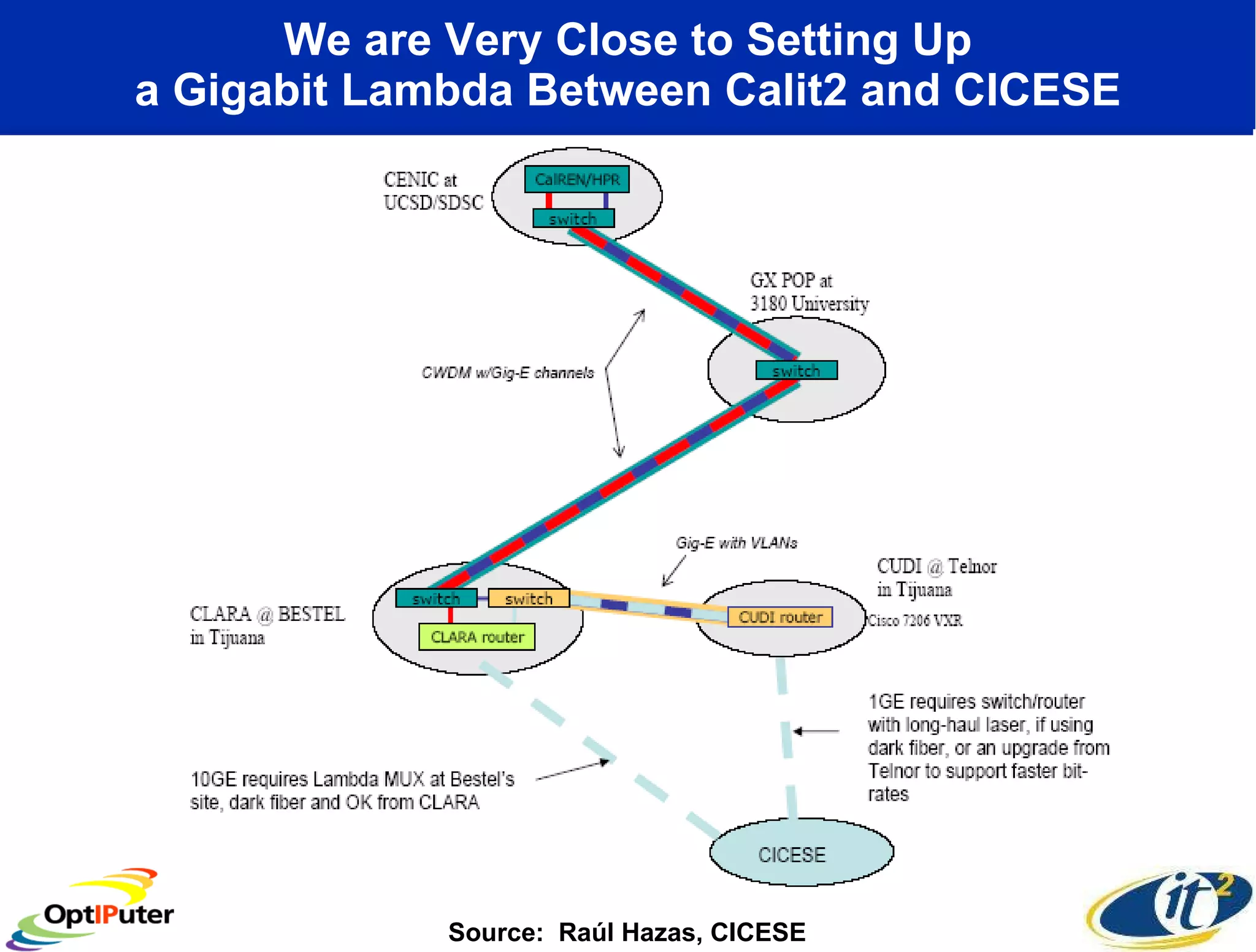
The document outlines the developments and initiatives of the CAMERA project, which focuses on cyberinfrastructure for advanced marine microbial ecology research. Led by Dr. Larry Smarr at UC San Diego, the project aims to enhance metagenomic data analysis and supports the sequencing and comparison of marine microbial genomes. It emphasizes the collaboration among various institutions and the integration of advanced networking technologies to facilitate global scientific collaboration in microbial genomics.