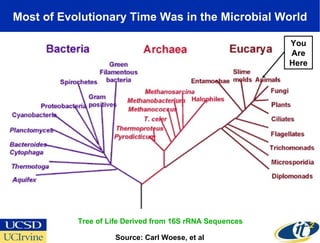
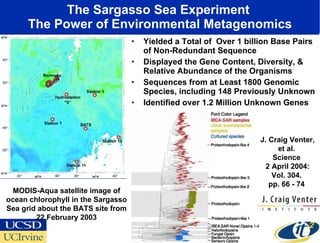
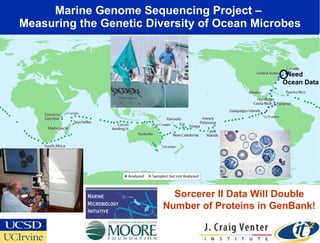
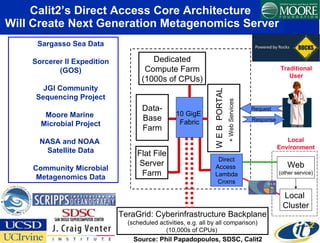
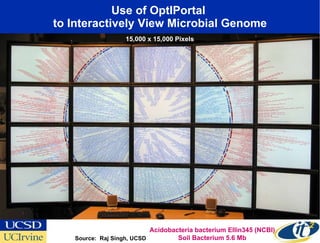
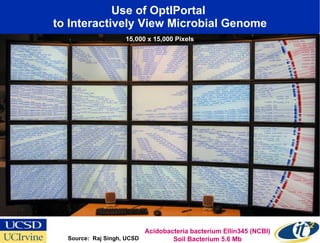
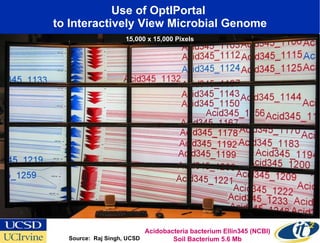
The Optiputer project, led by Dr. Larry Smarr, aims to enhance metagenomics research through high-resolution data portals over dedicated optical channels, facilitating collaboration among global researchers. With a significant investment of $13.5 million over five years, the project has generated extensive genetic data, including over 1.2 million unknown genes from ocean microbes. The initiative integrates advanced computing infrastructure to enable interactive genome visualization and foster scientific discovery in microbial diversity and environmental genomics.