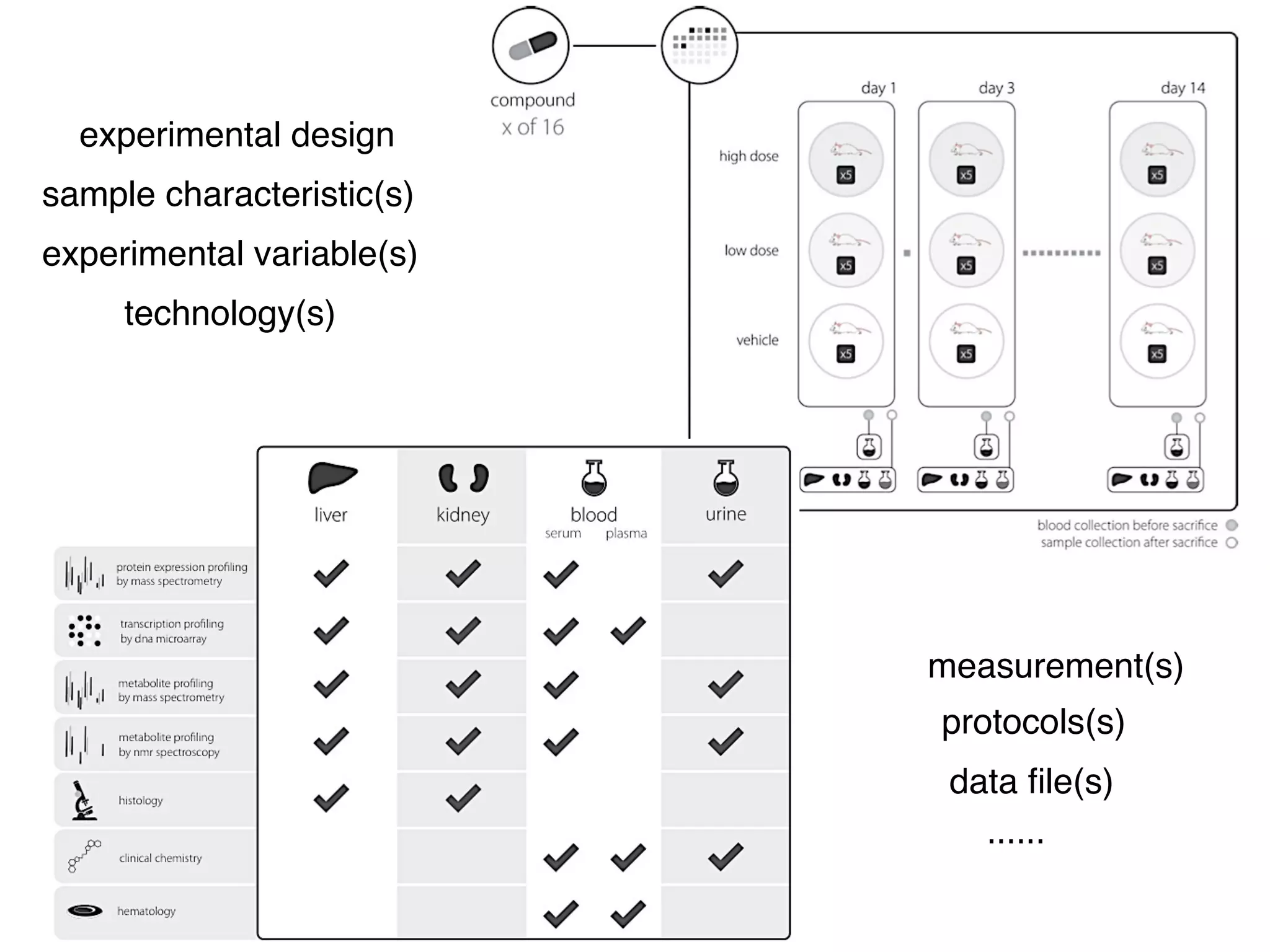
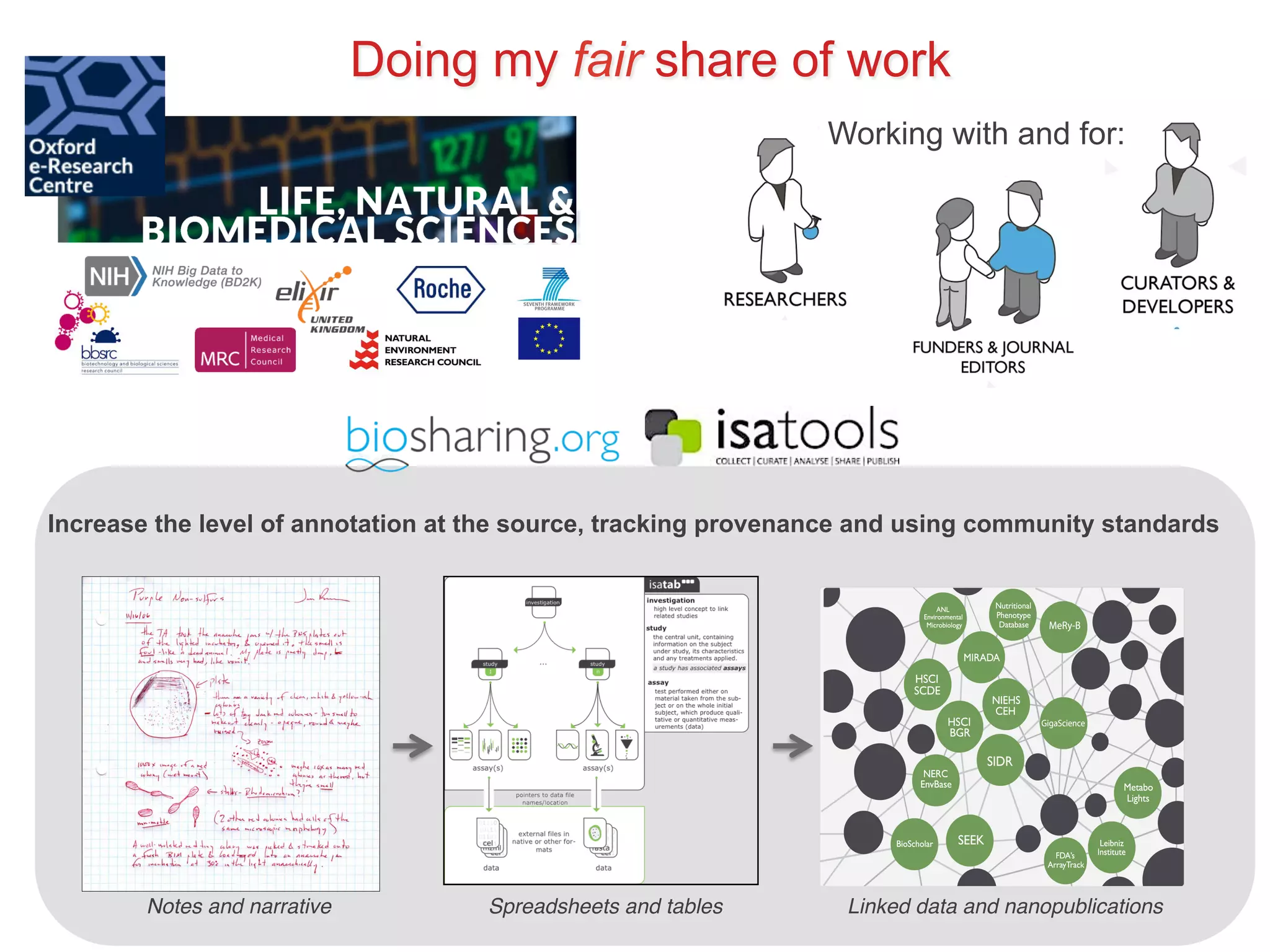
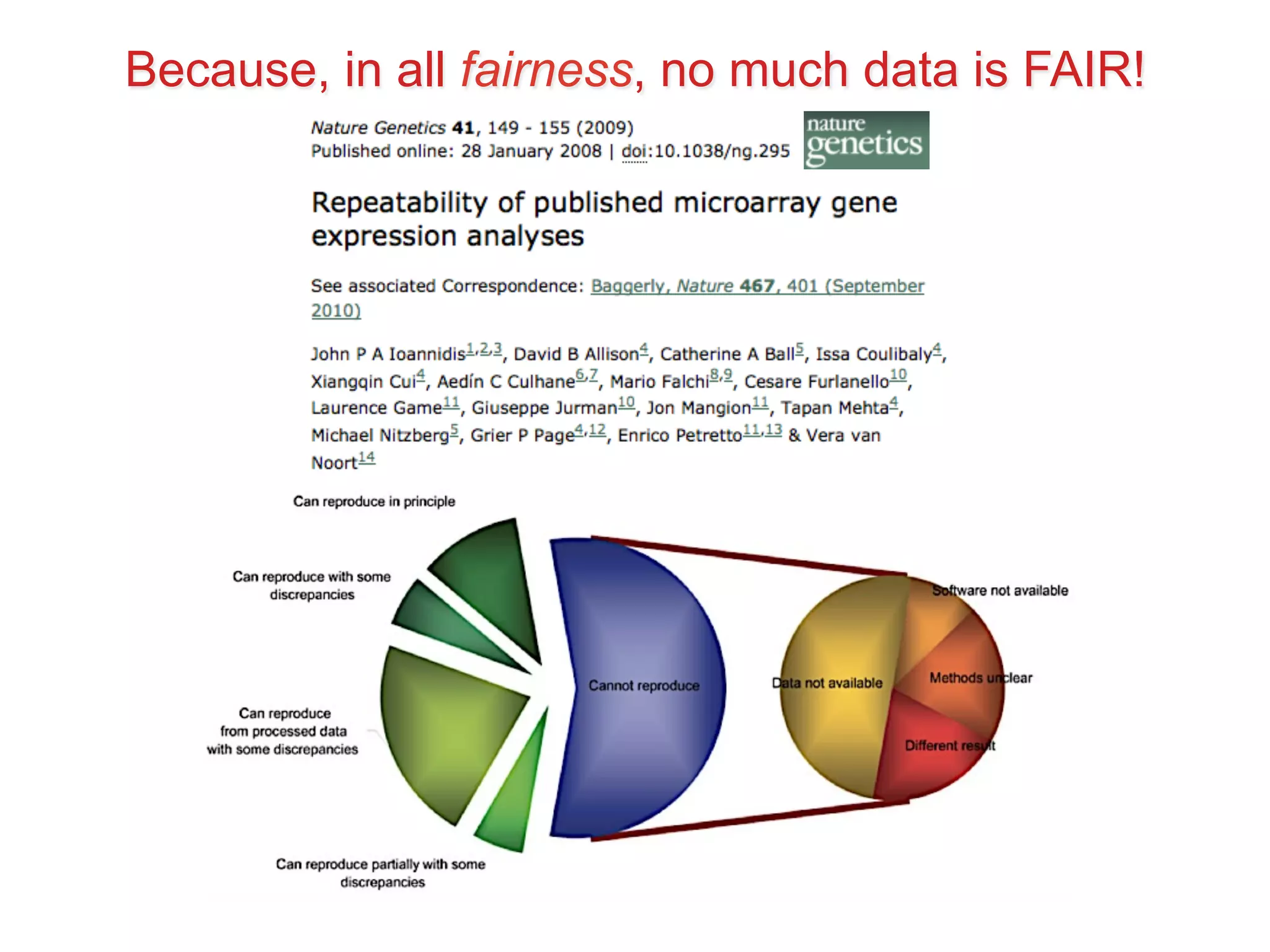
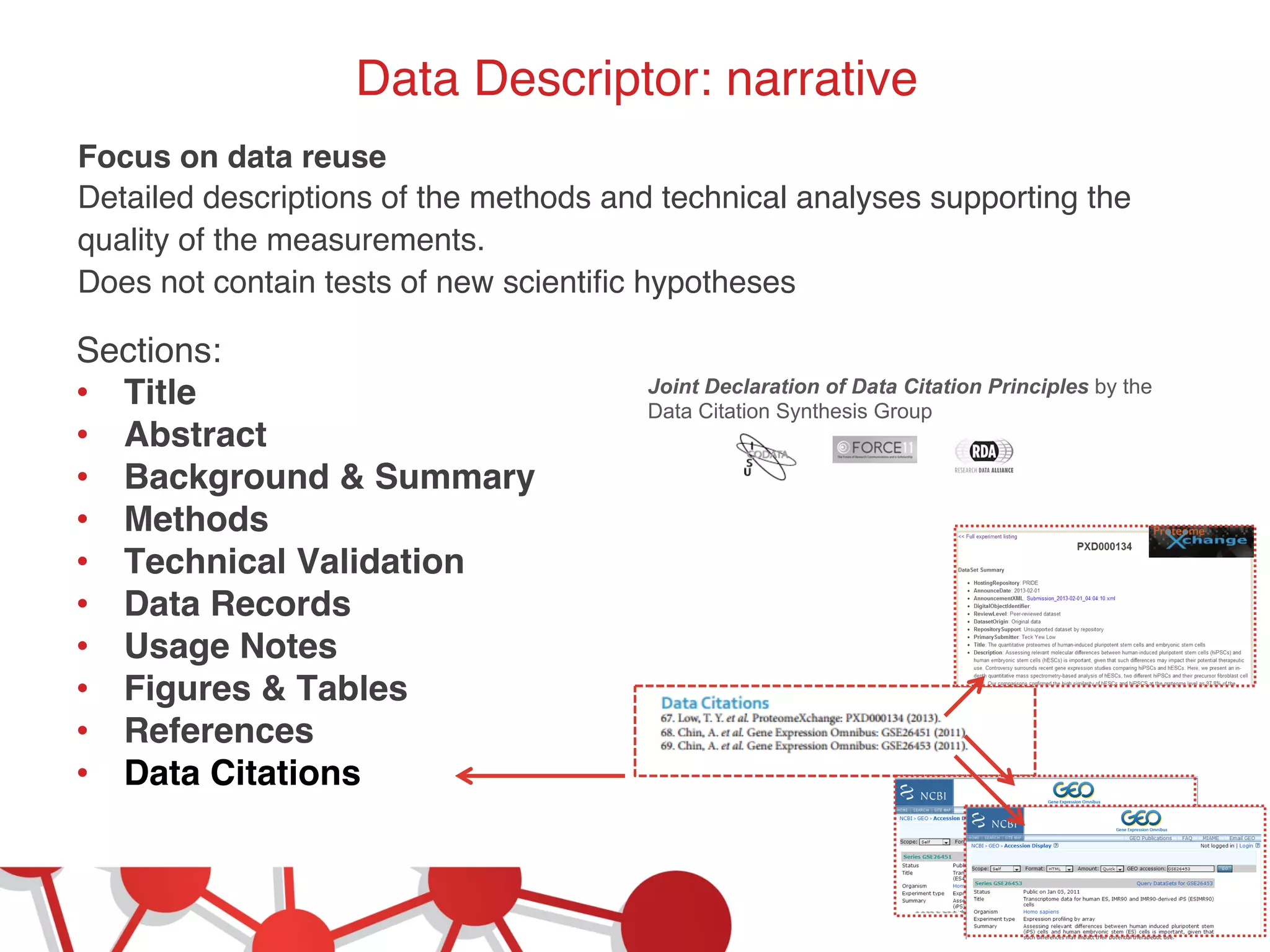
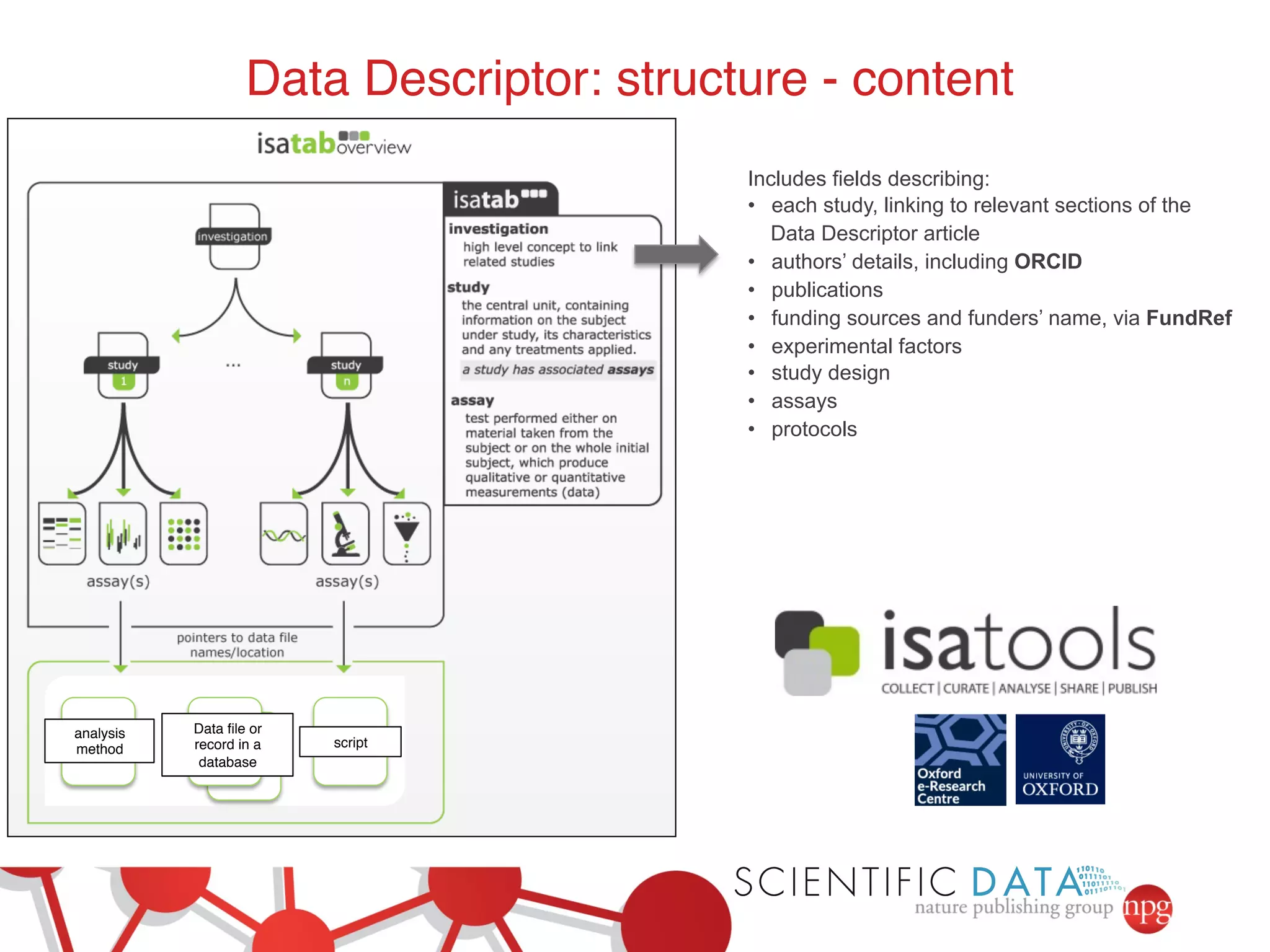
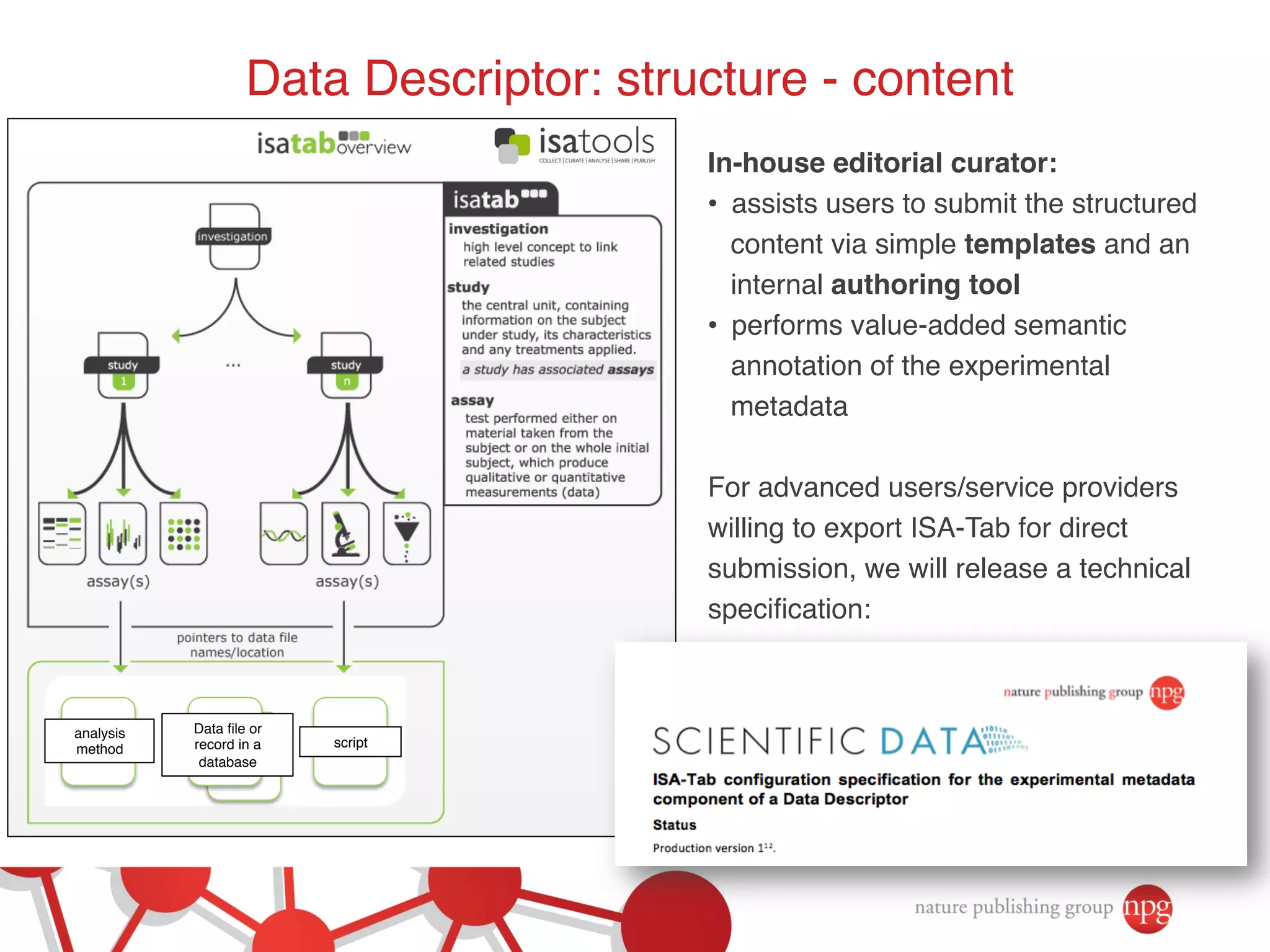
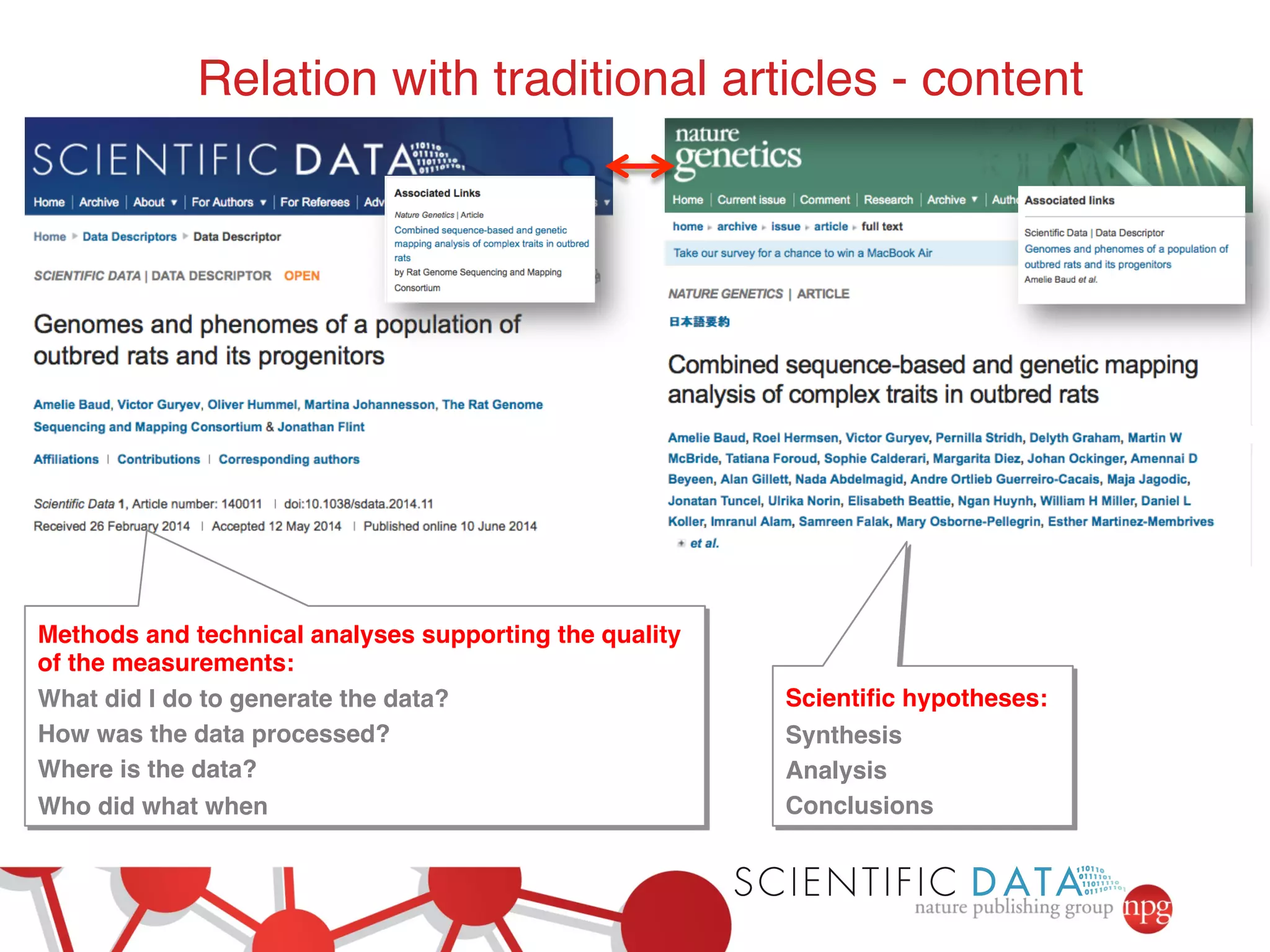
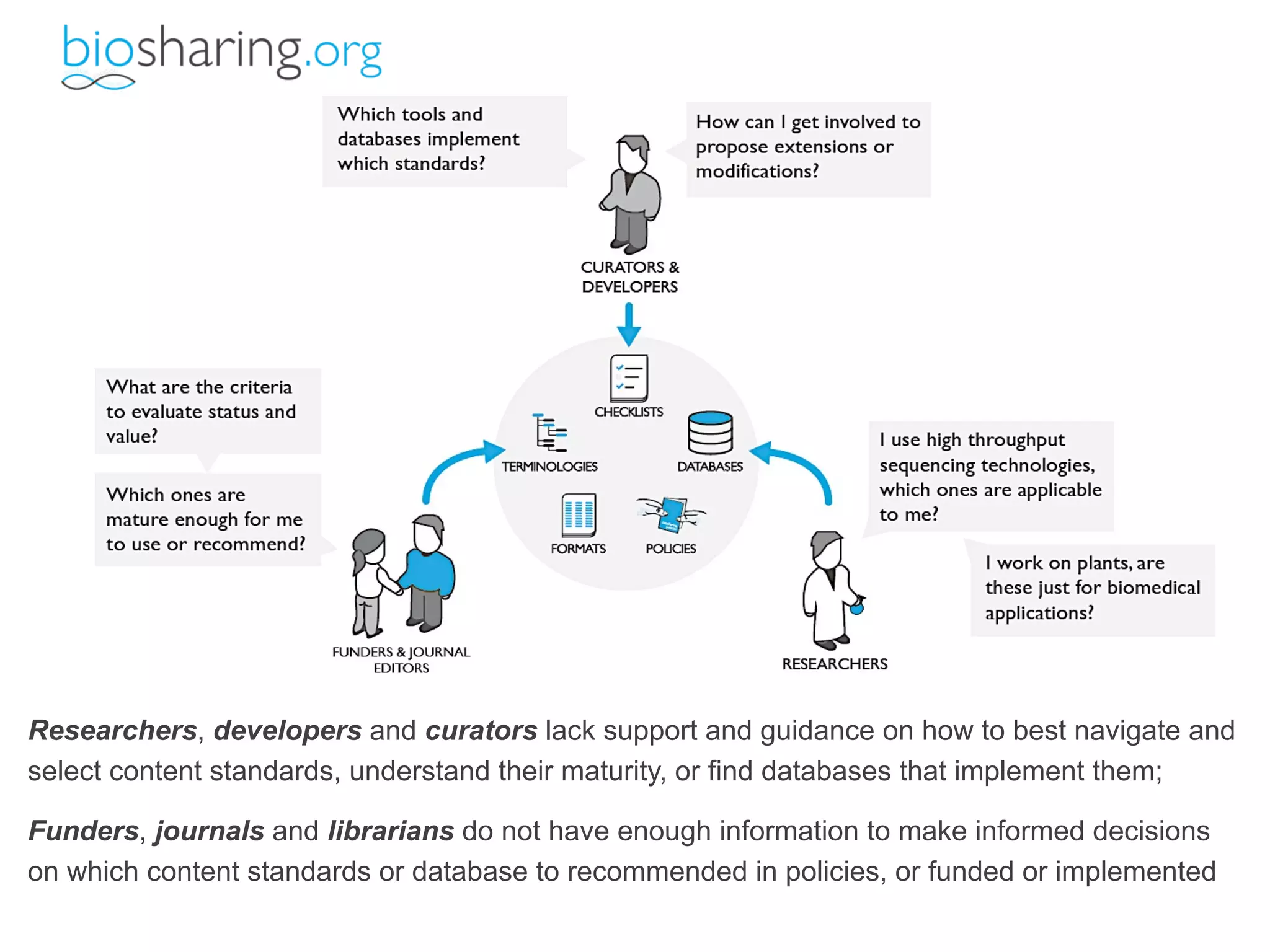
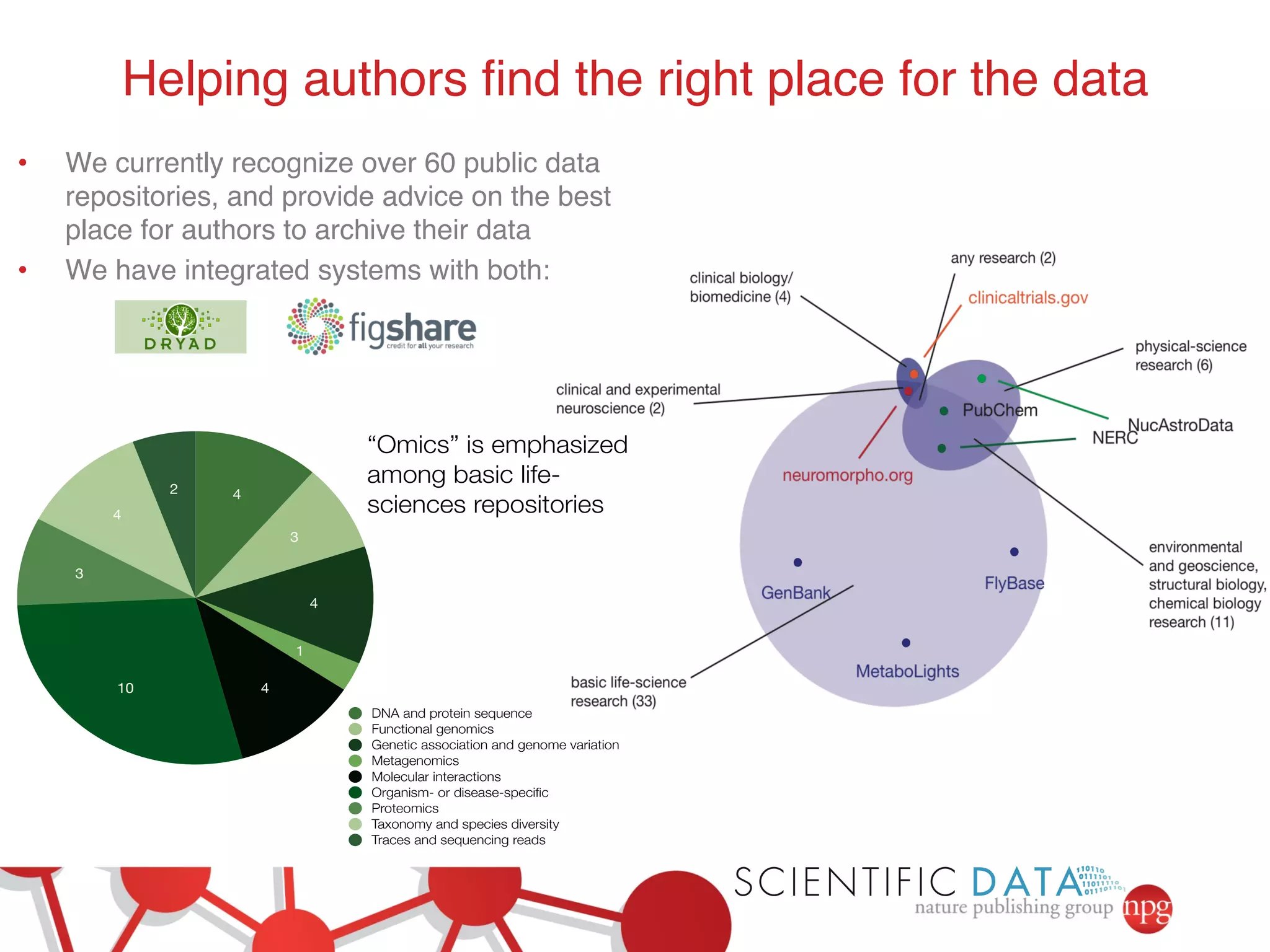
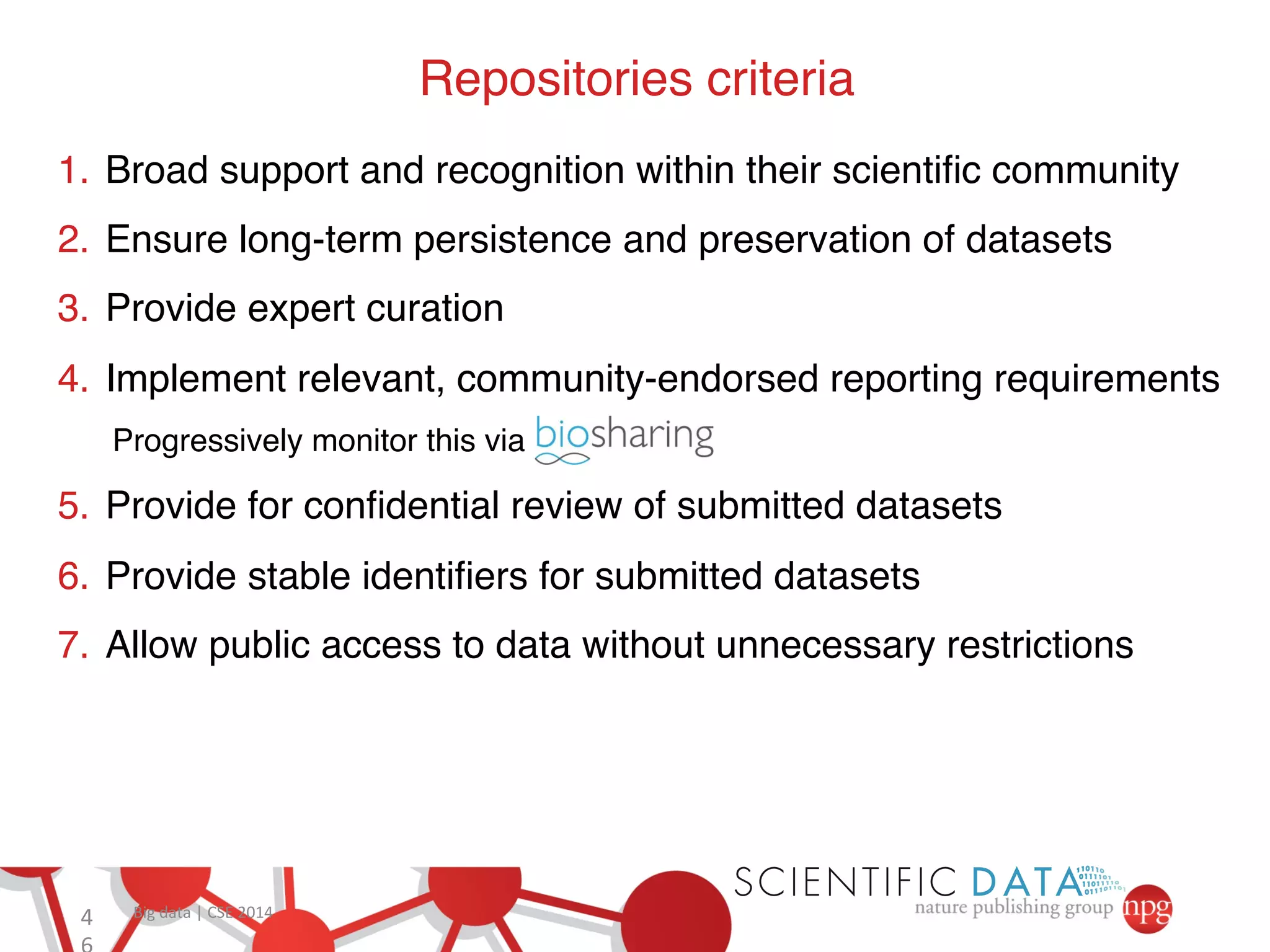
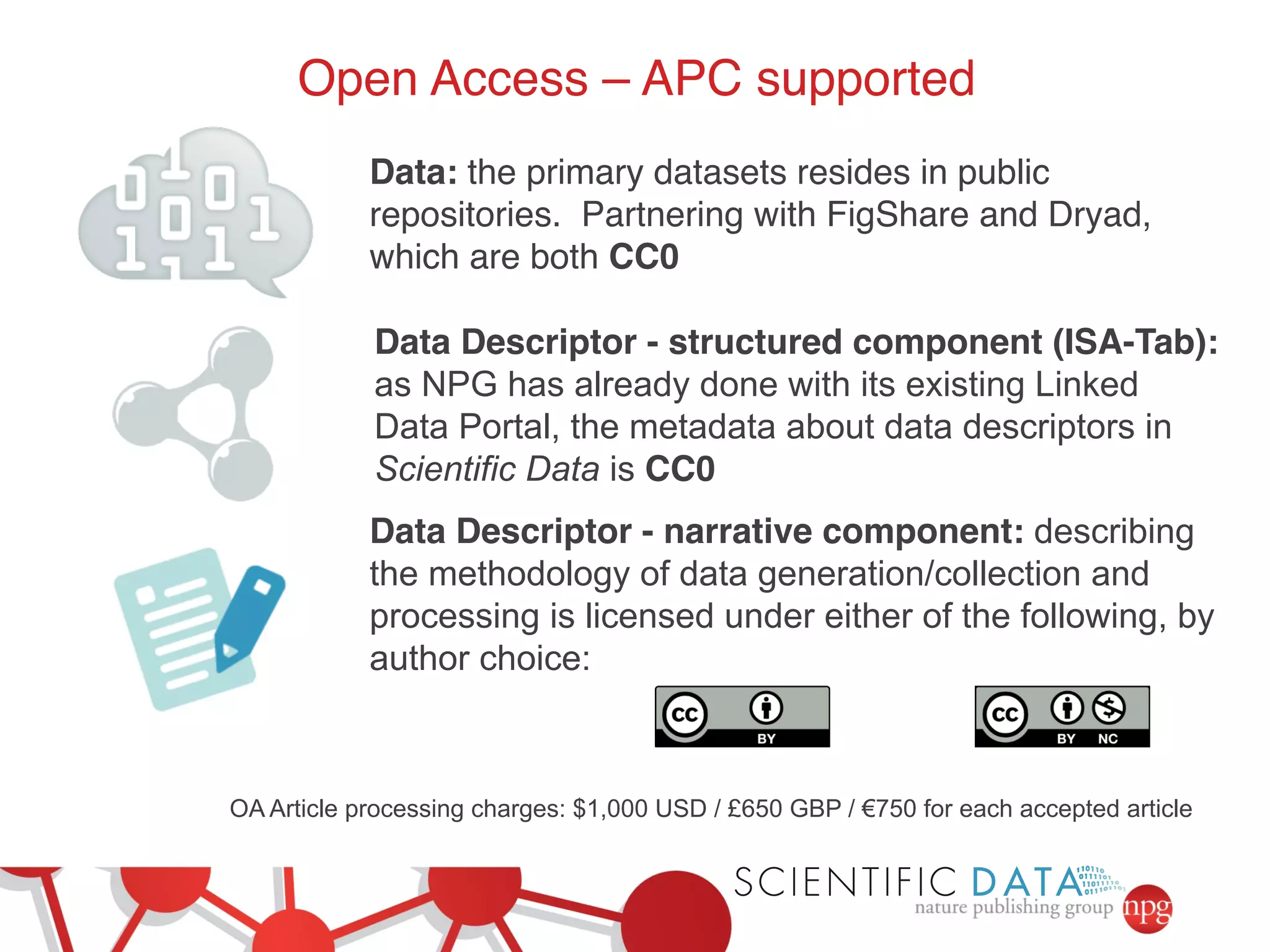
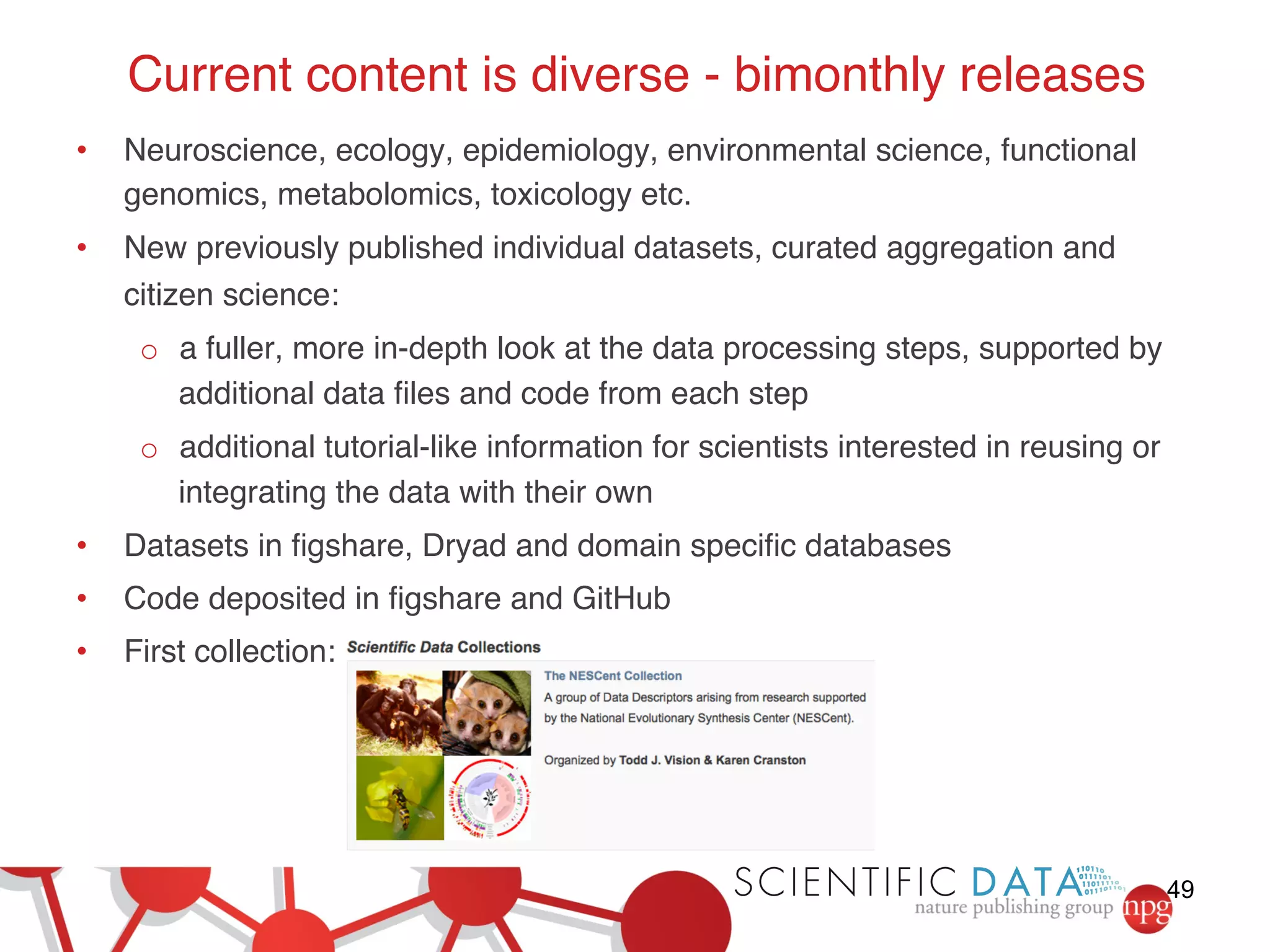
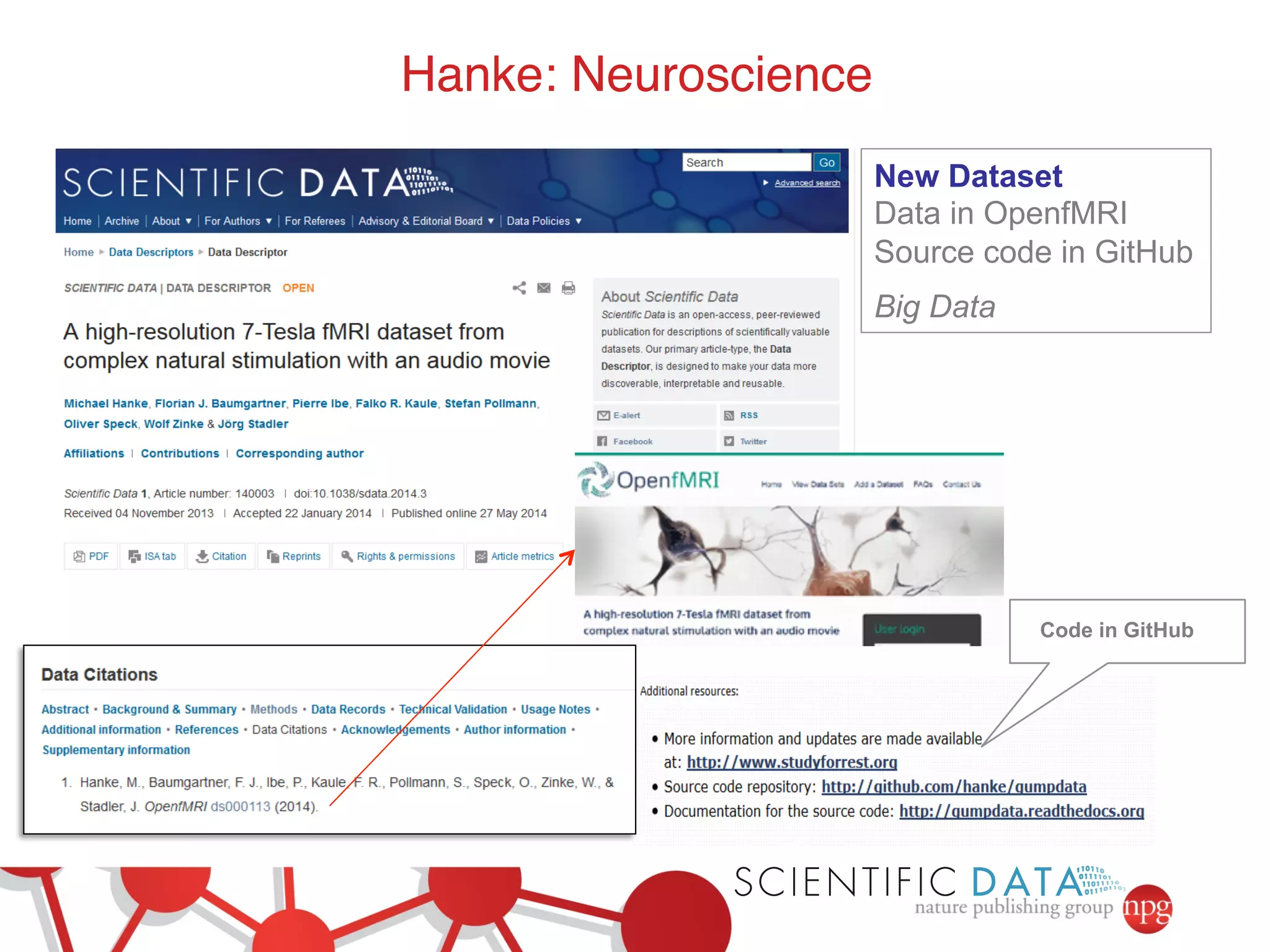
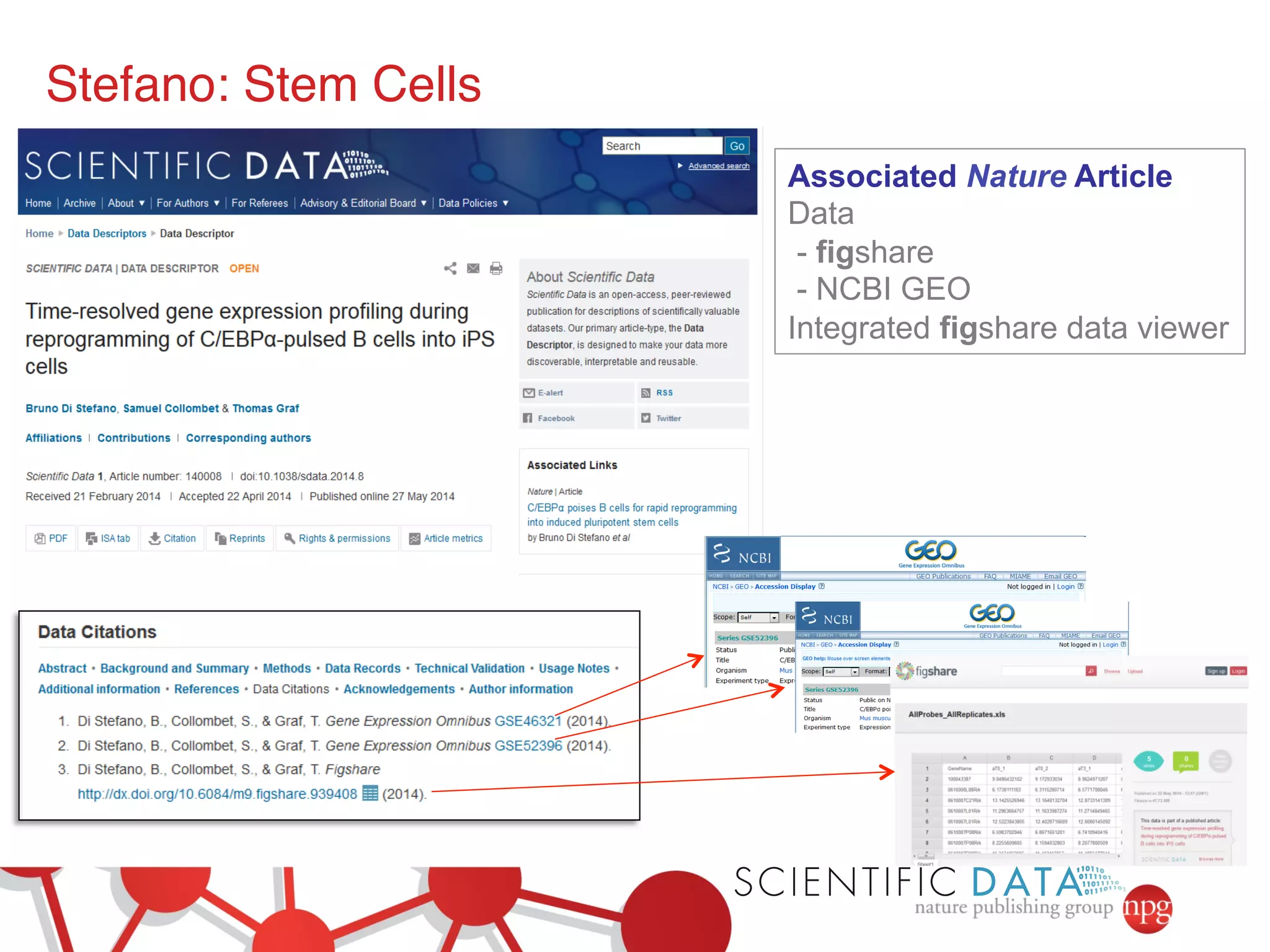
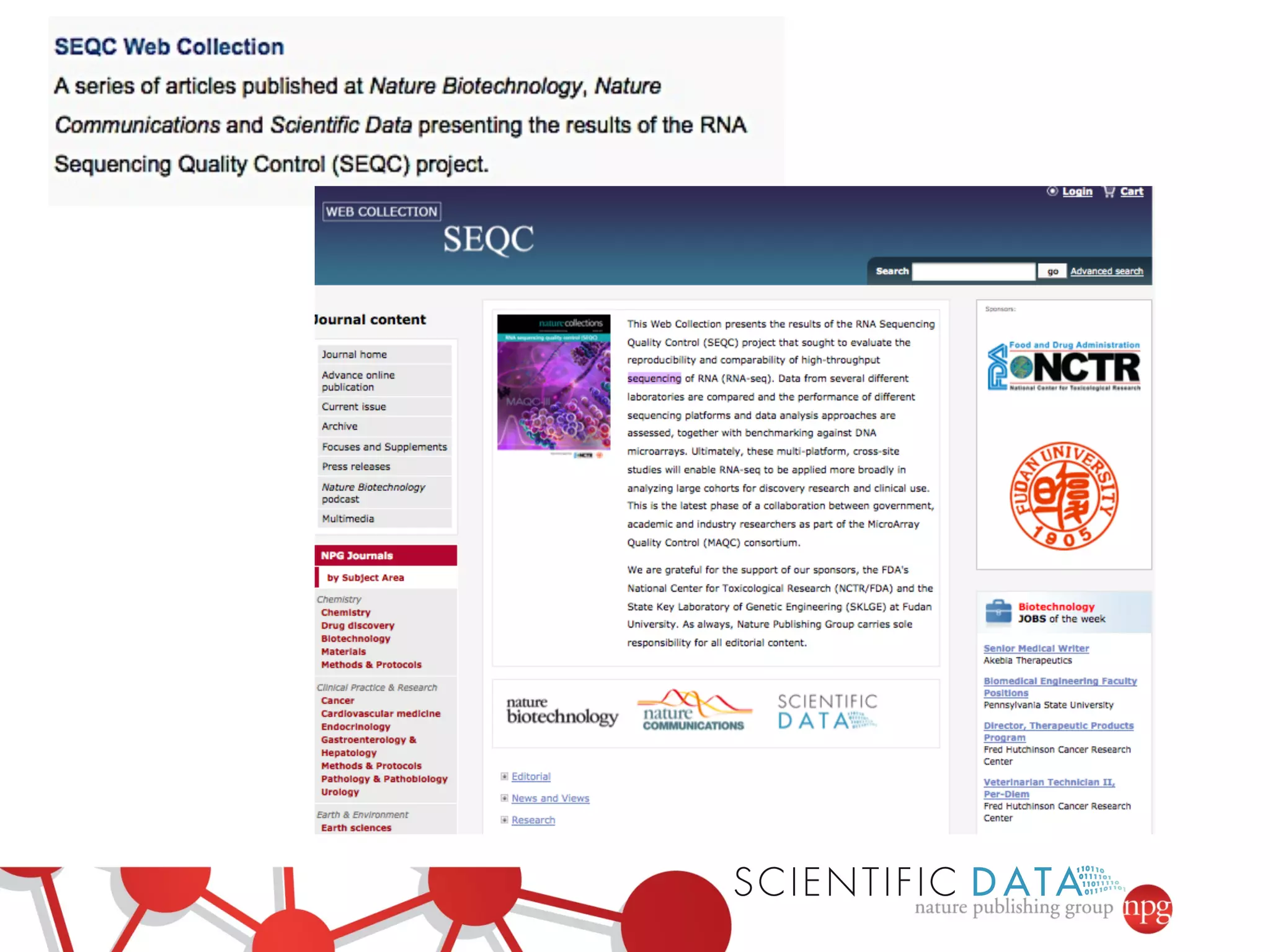
This document summarizes Susanna-Assunta Sansone's presentation on open access and open data at Nature Publishing Group. Some key points discussed include:
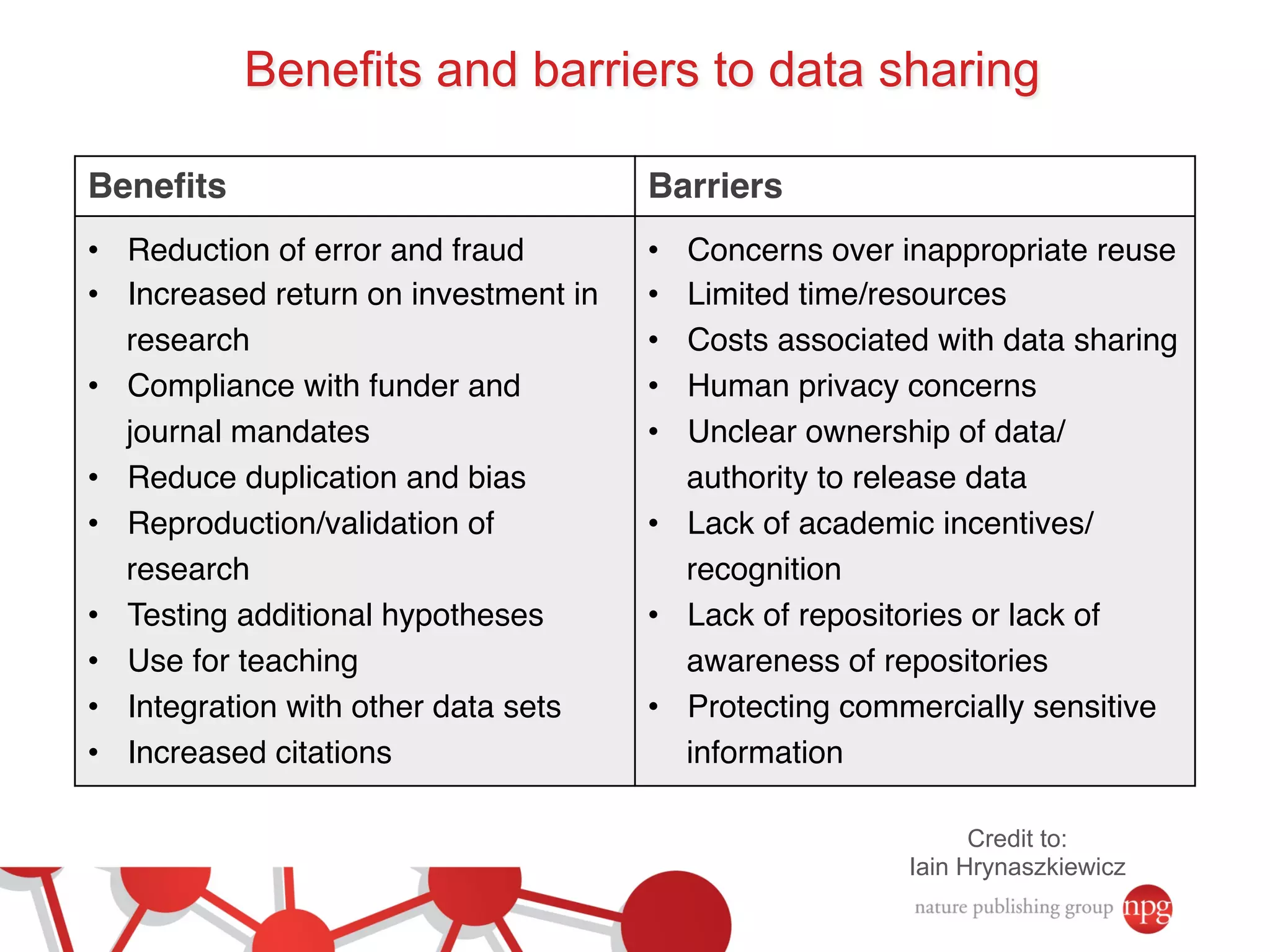
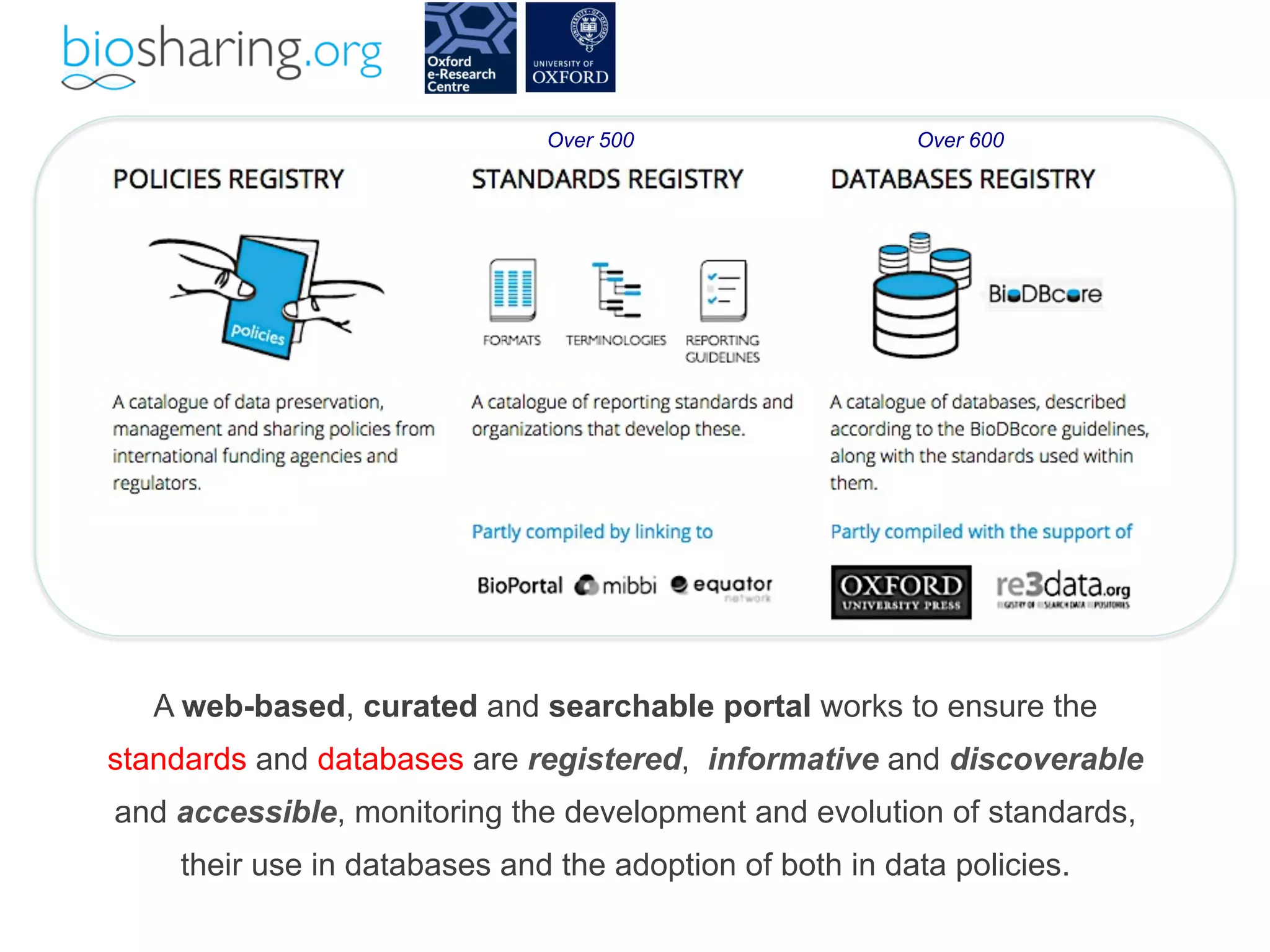
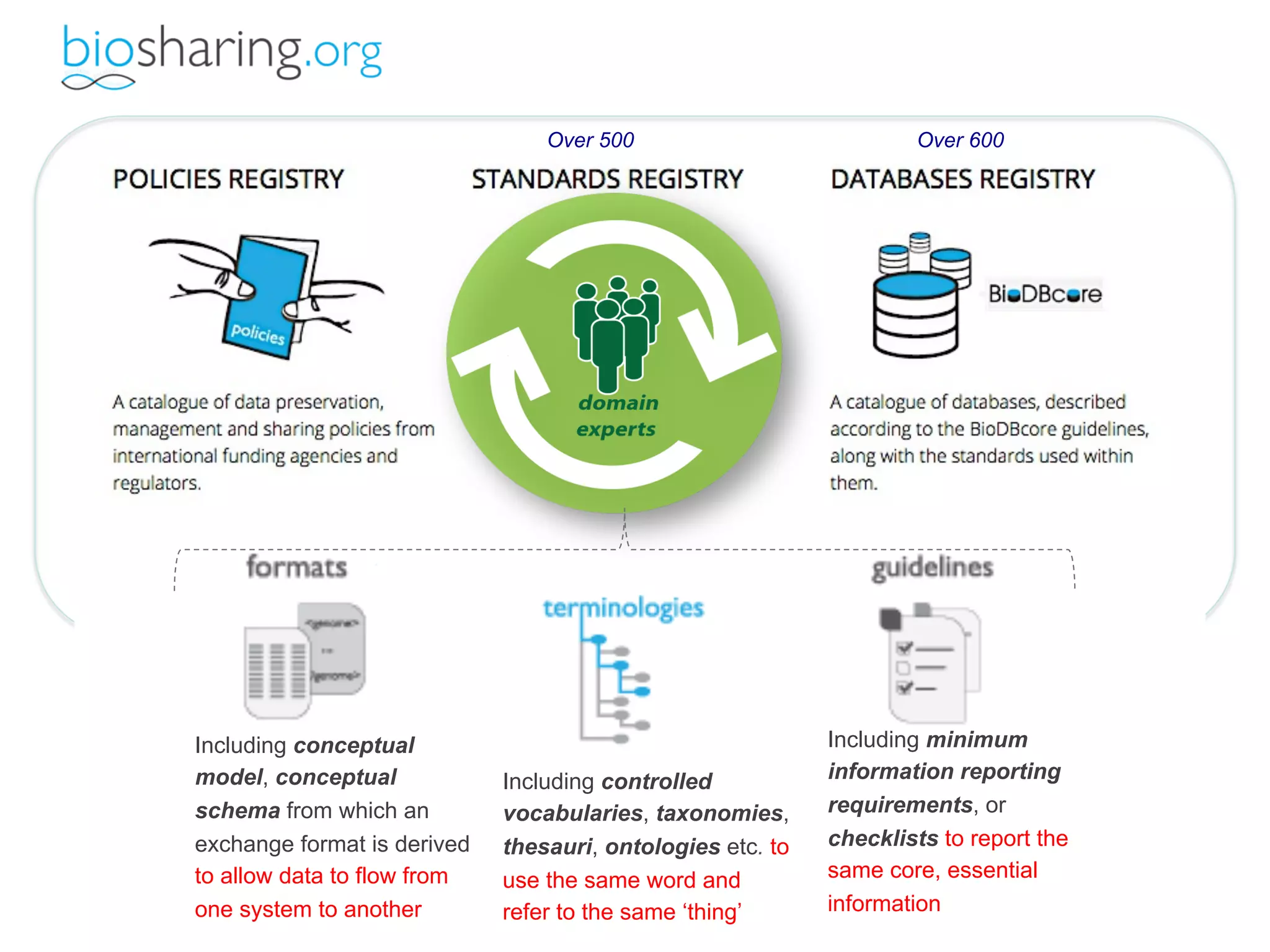
- The benefits of open data including reducing errors/fraud and increasing return on investment in research. However, barriers also exist such as lack of incentives and standards.
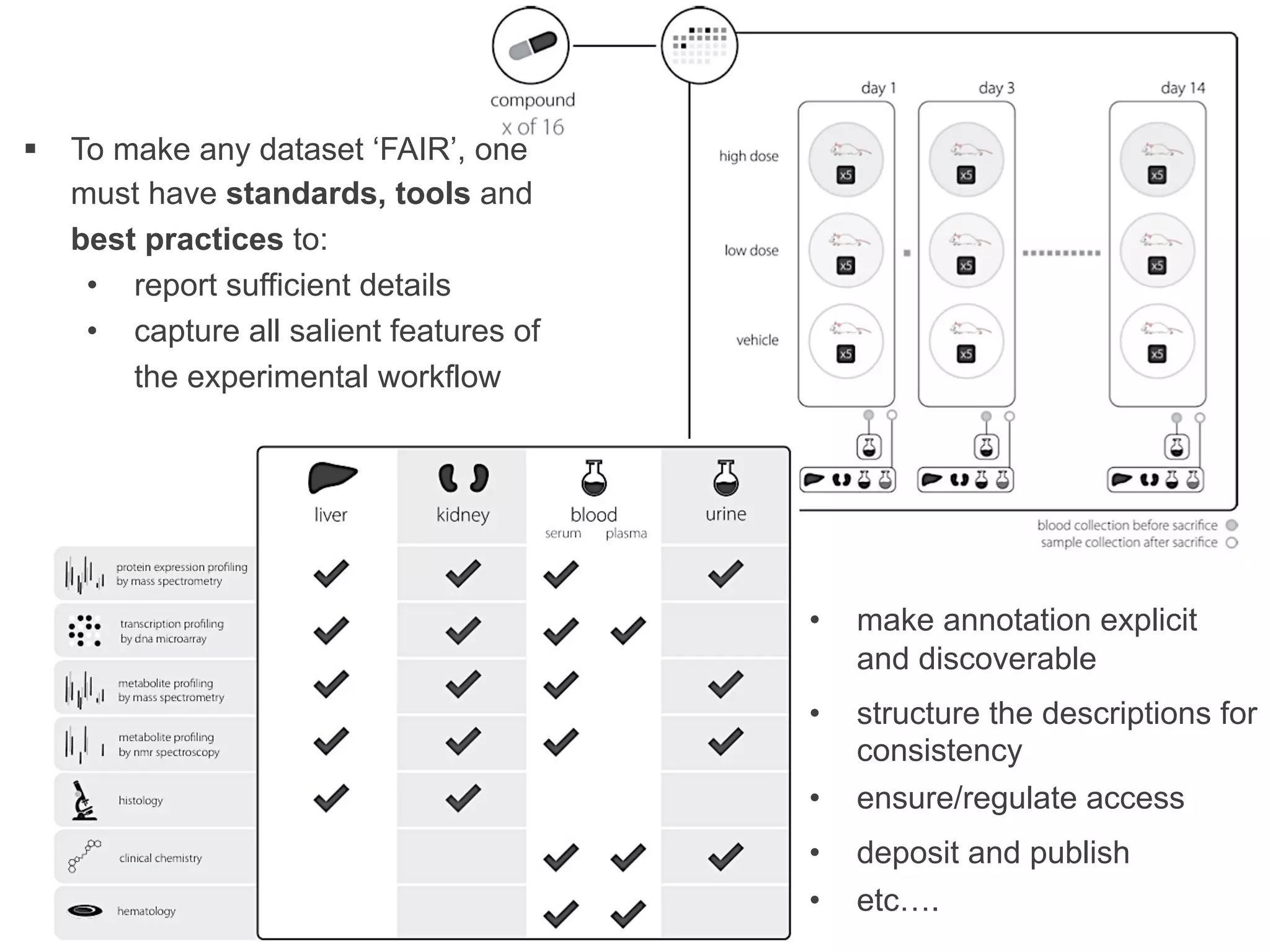
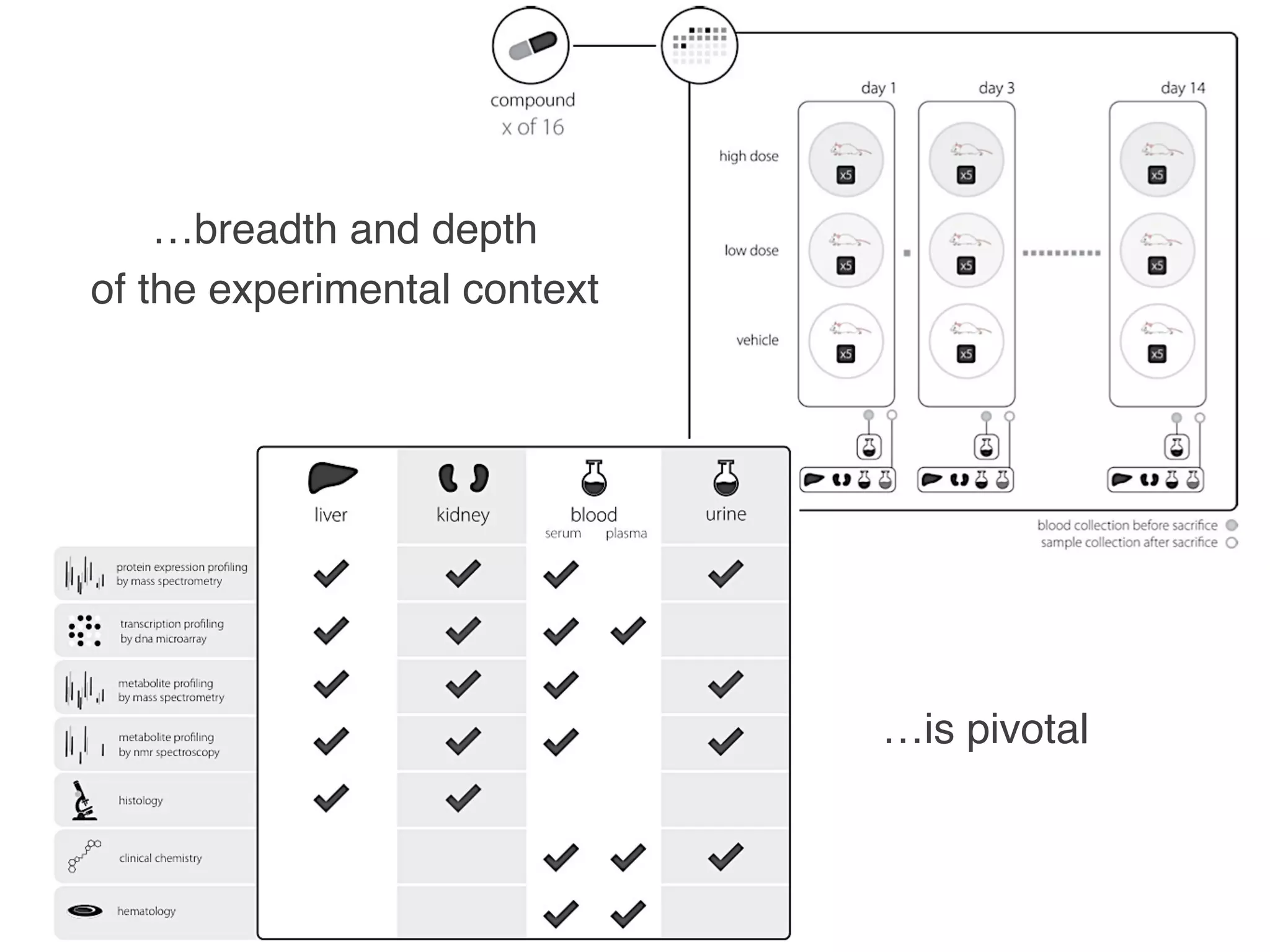
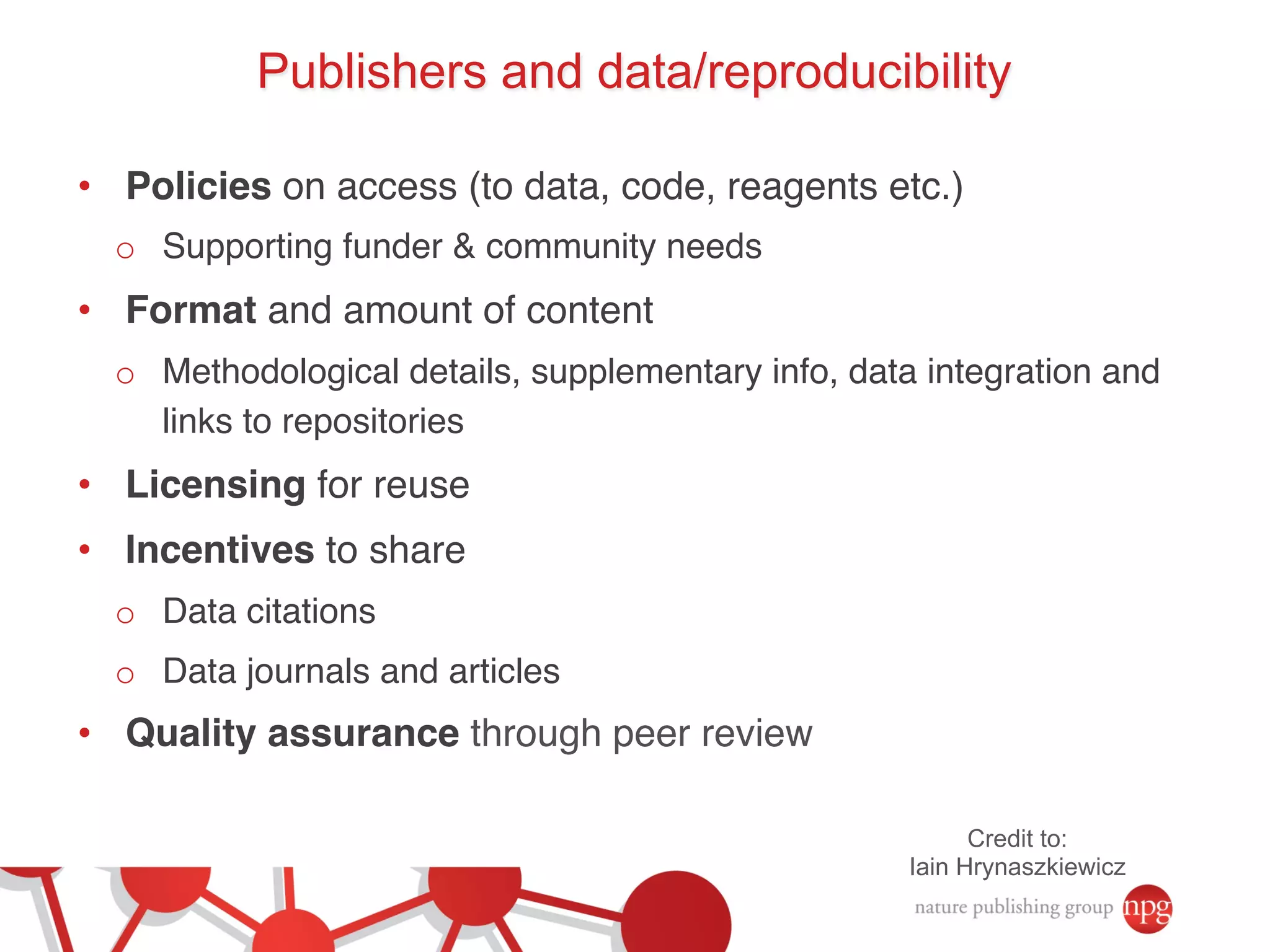
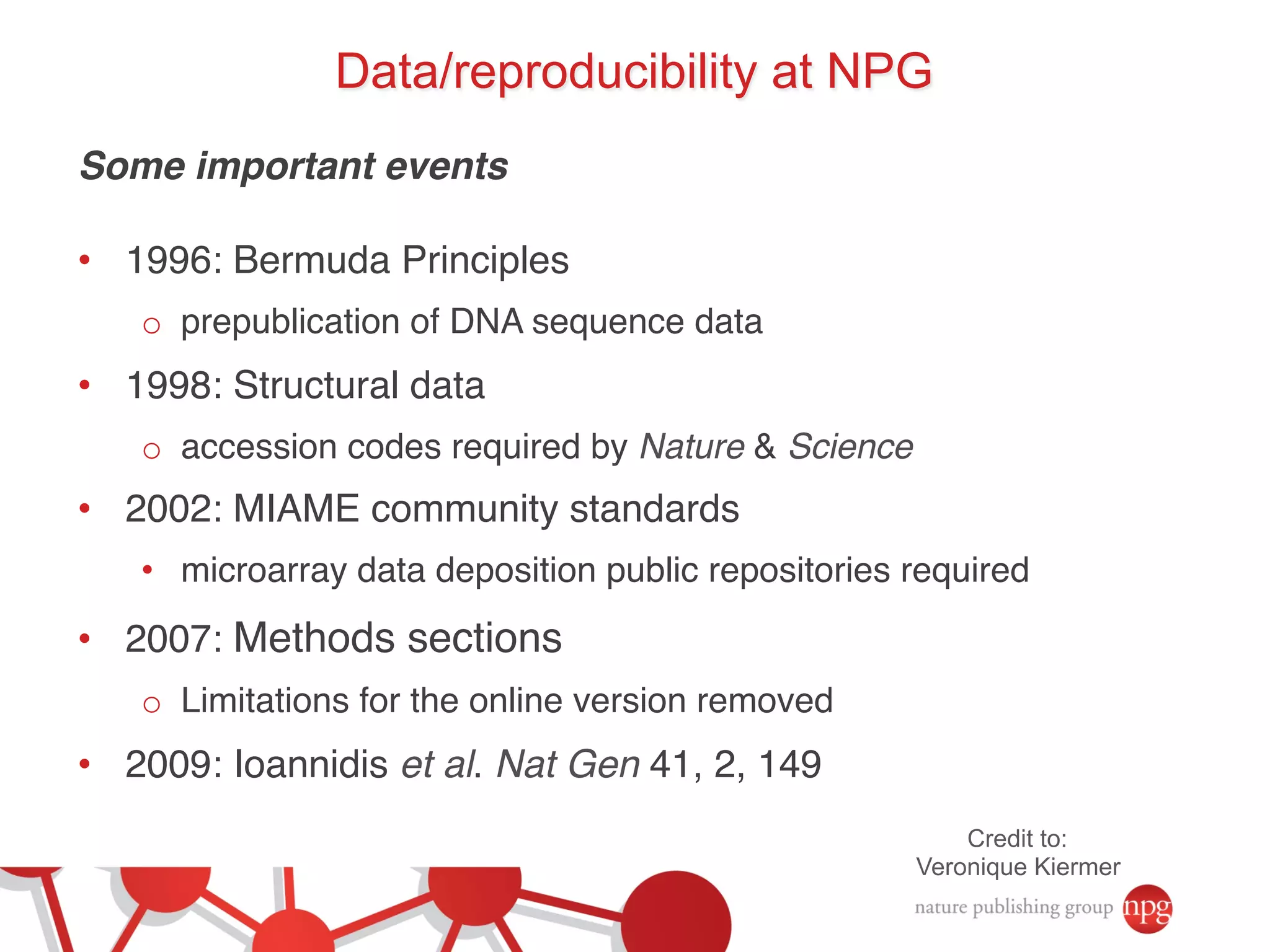
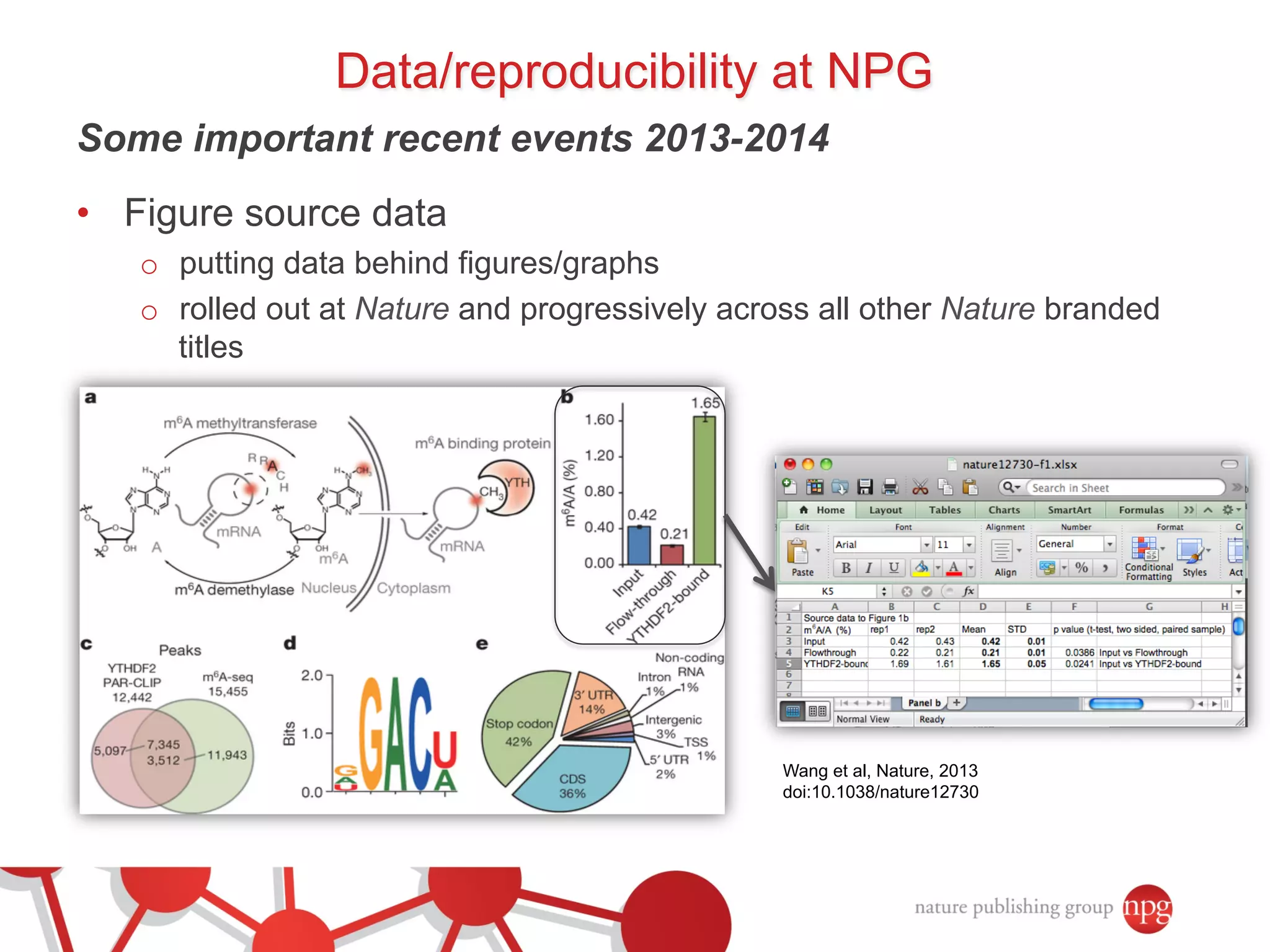
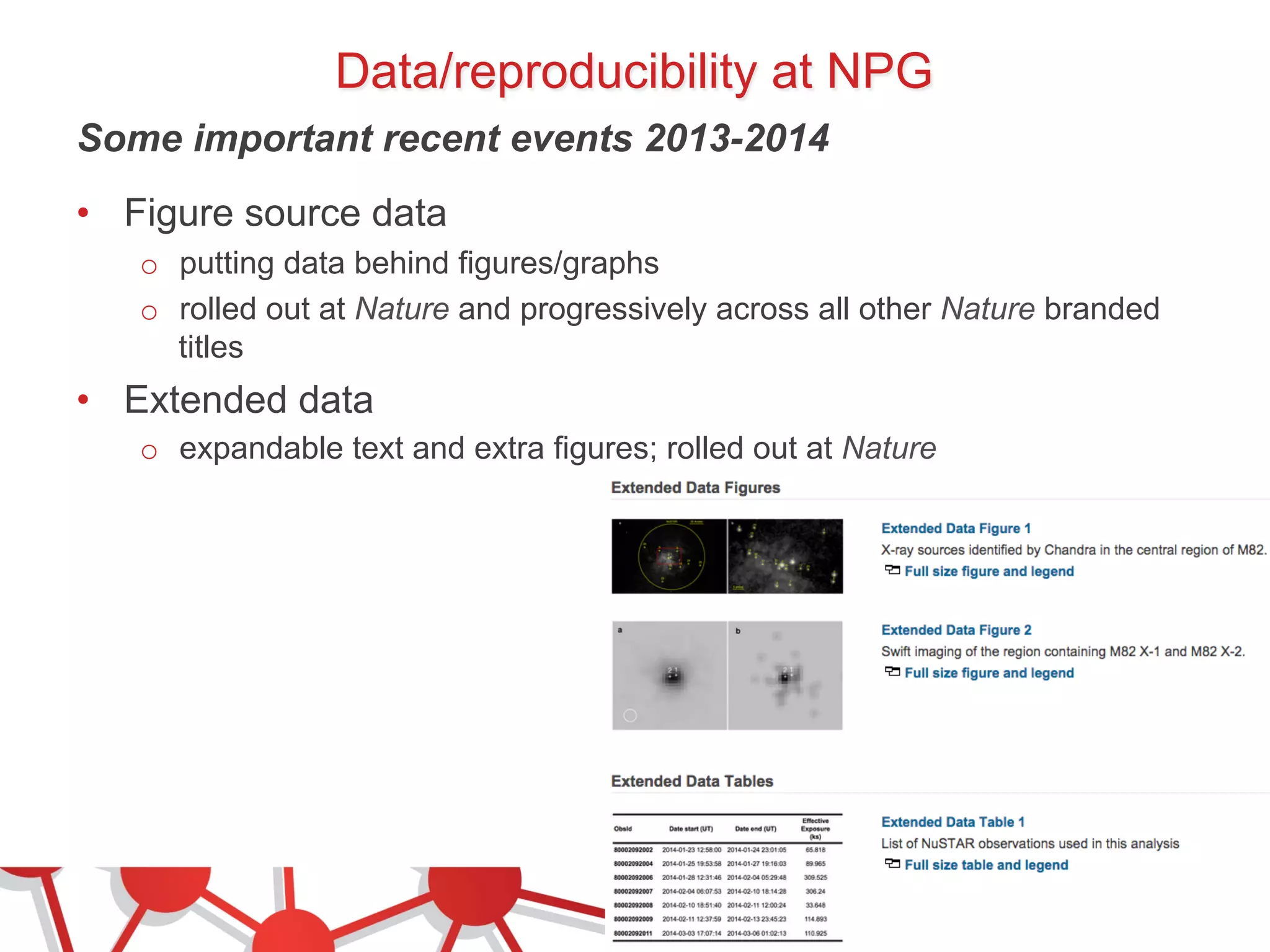
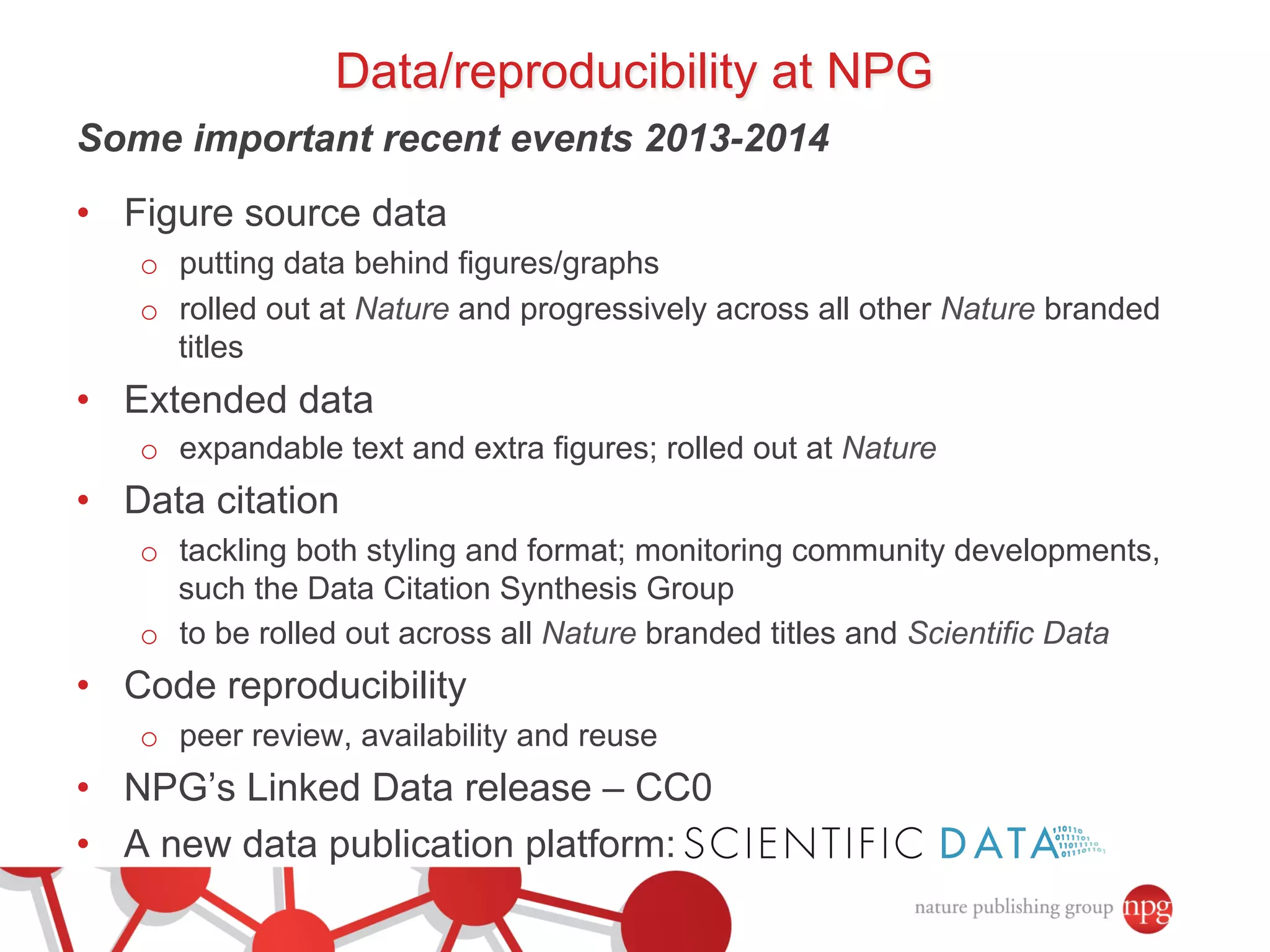
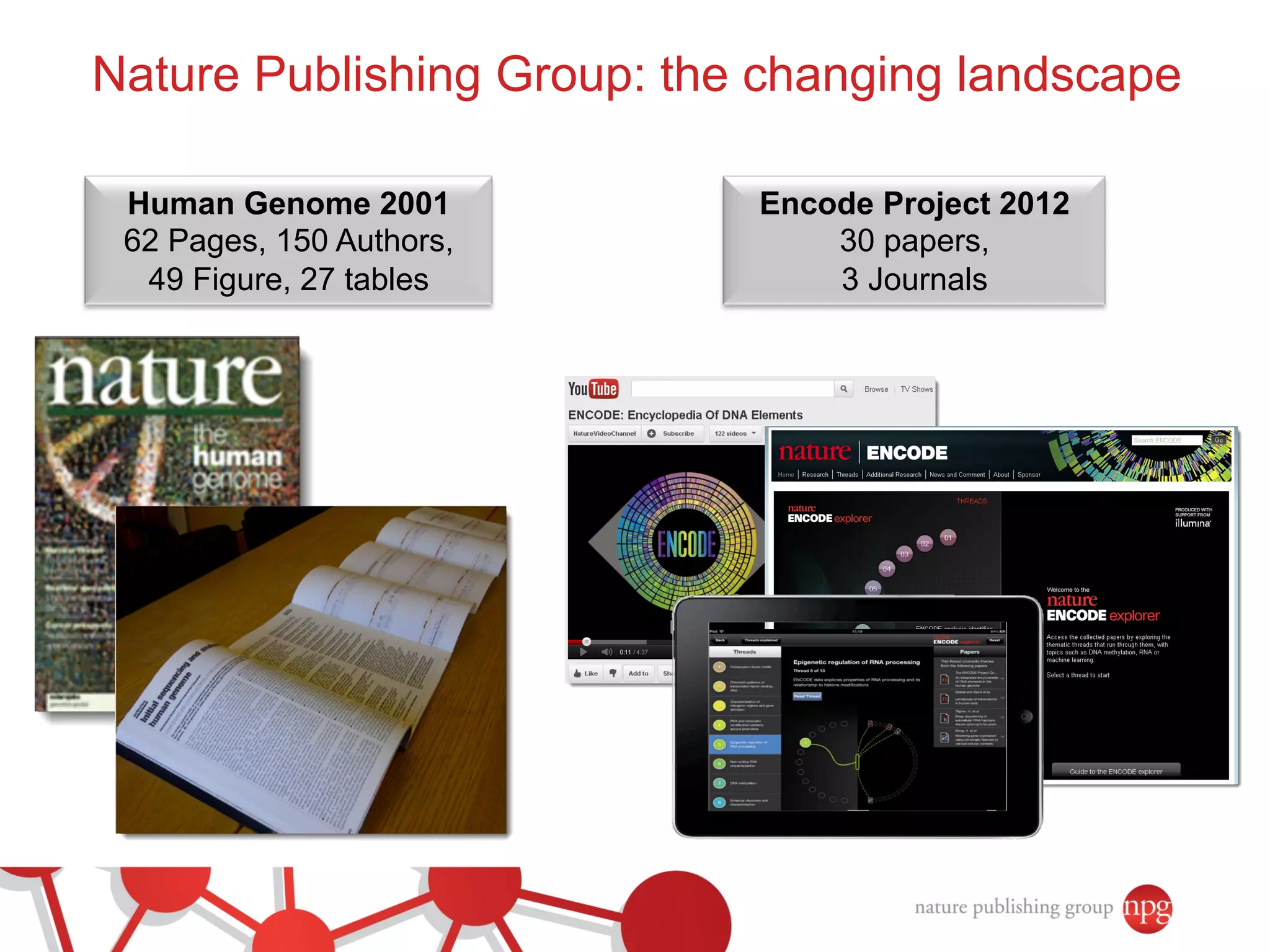
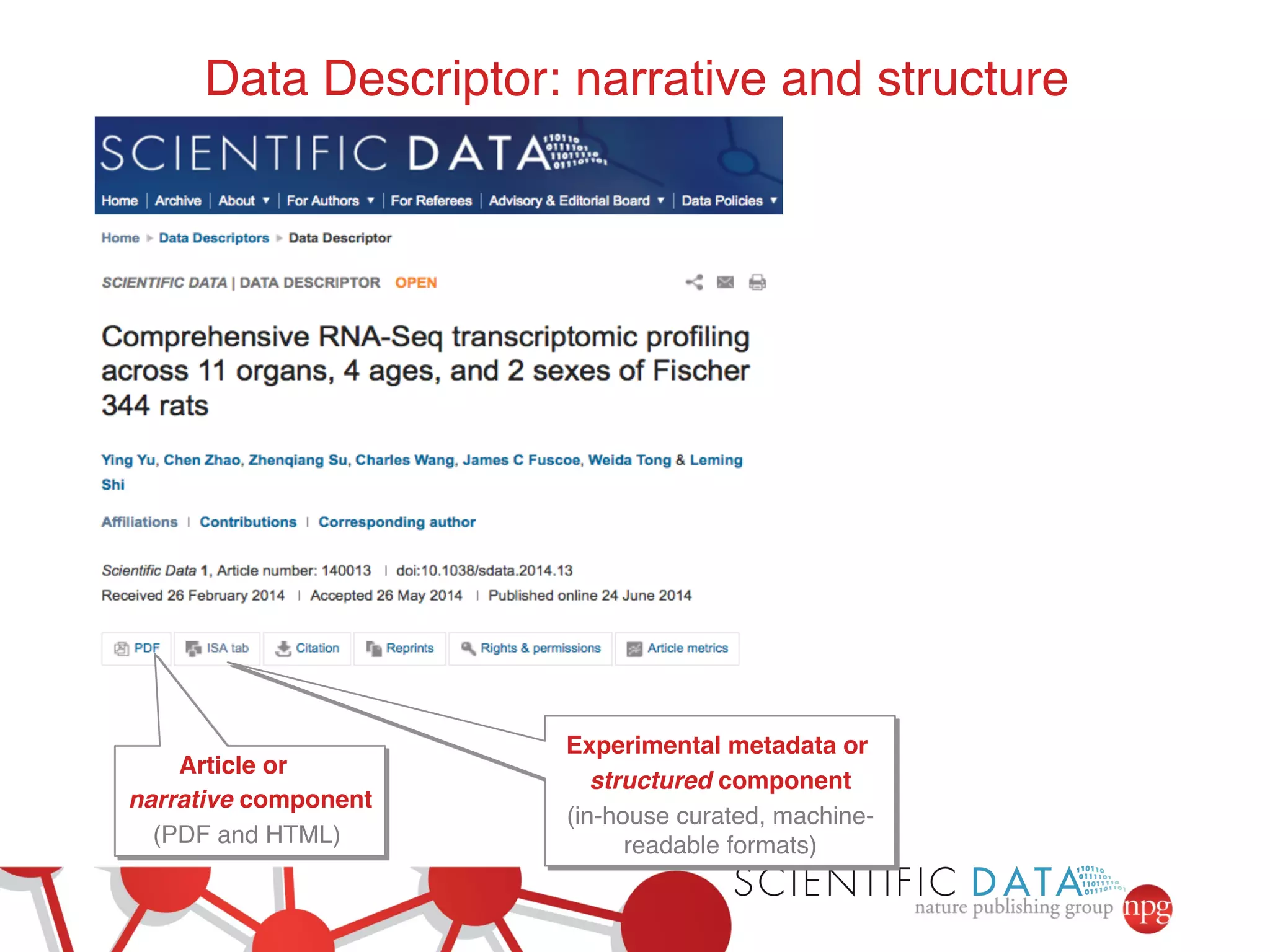
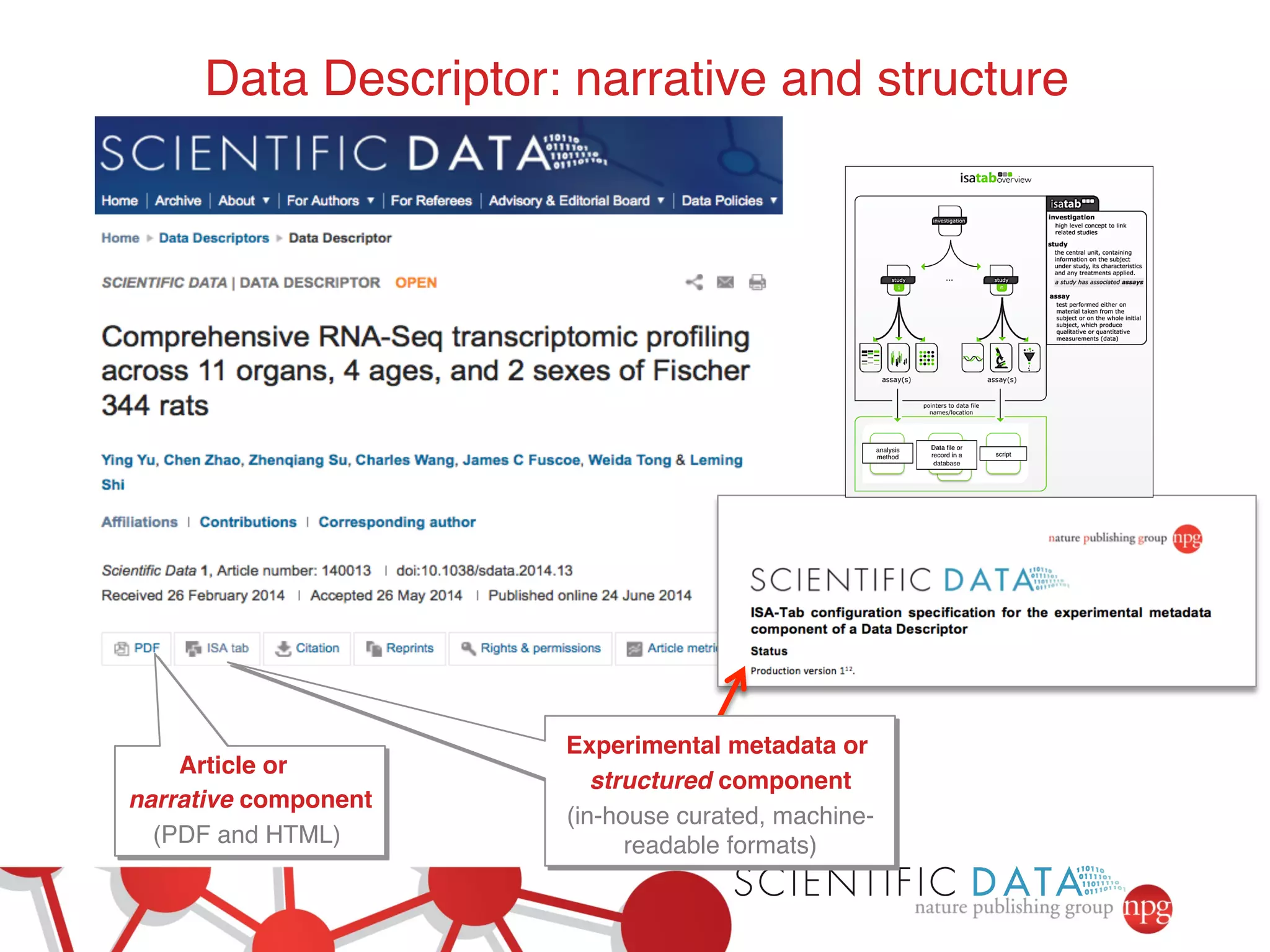
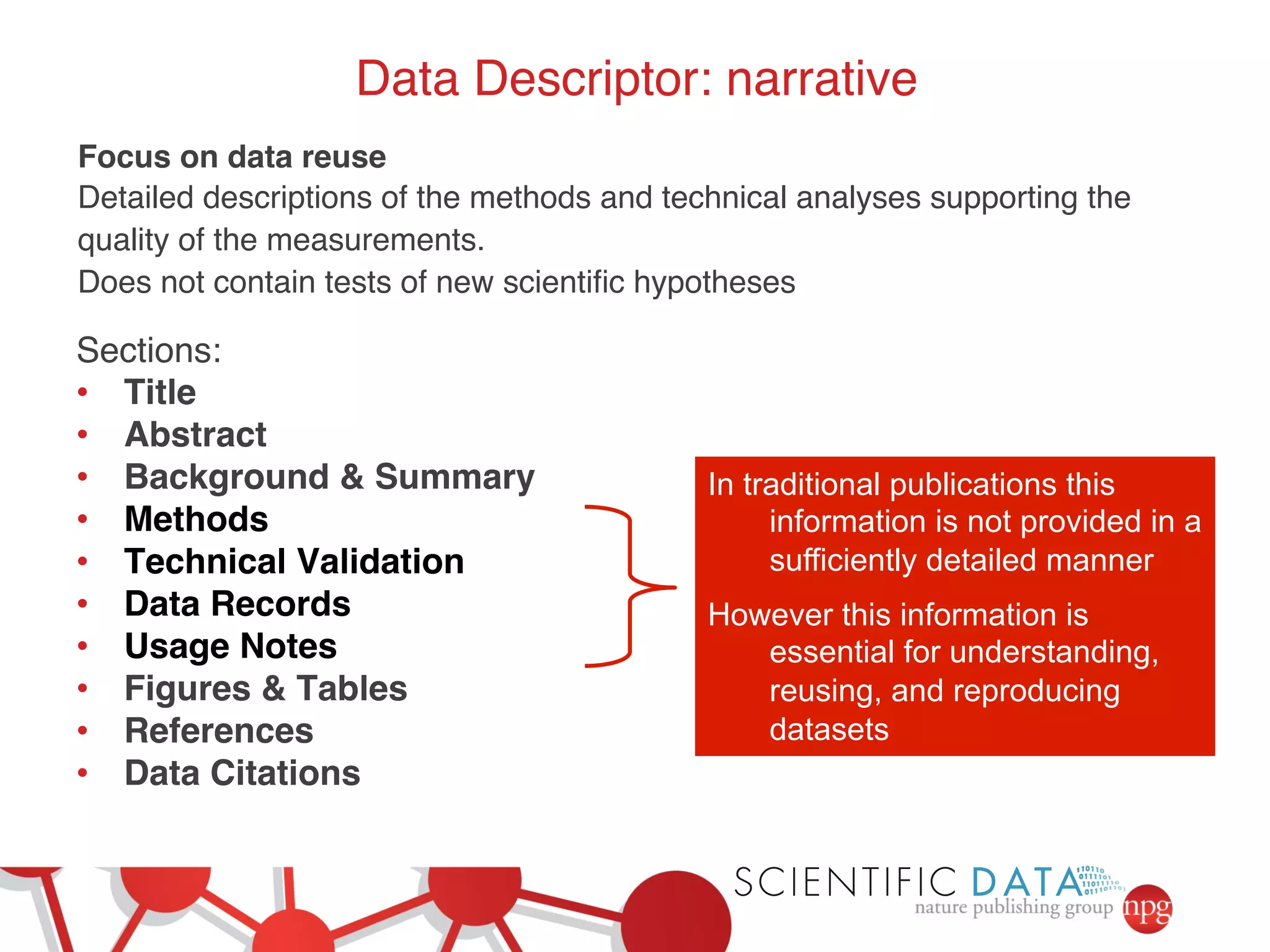
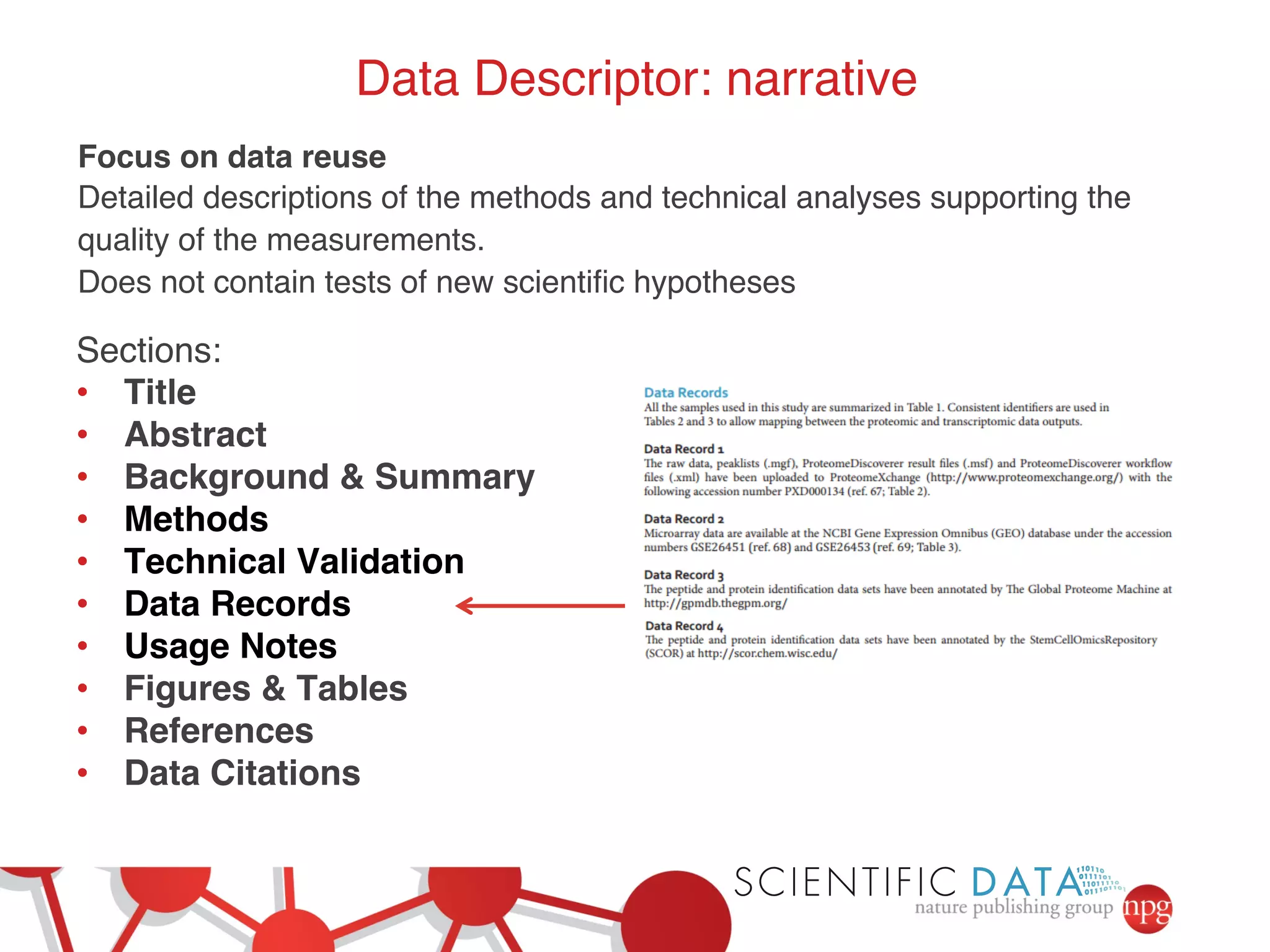
- Recent initiatives at NPG to improve data/reproducibility such as requiring data behind figures and expanding methods sections.
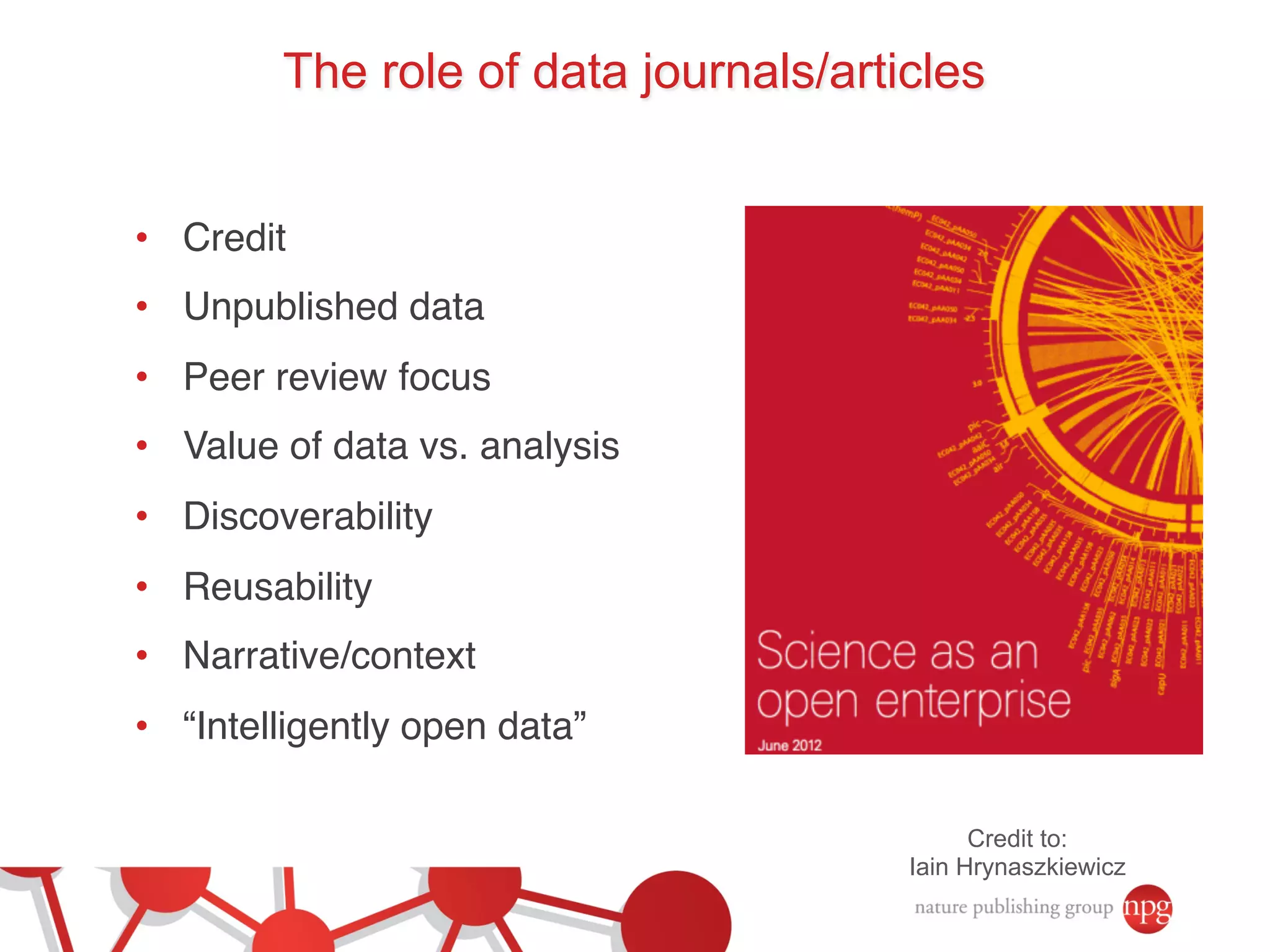
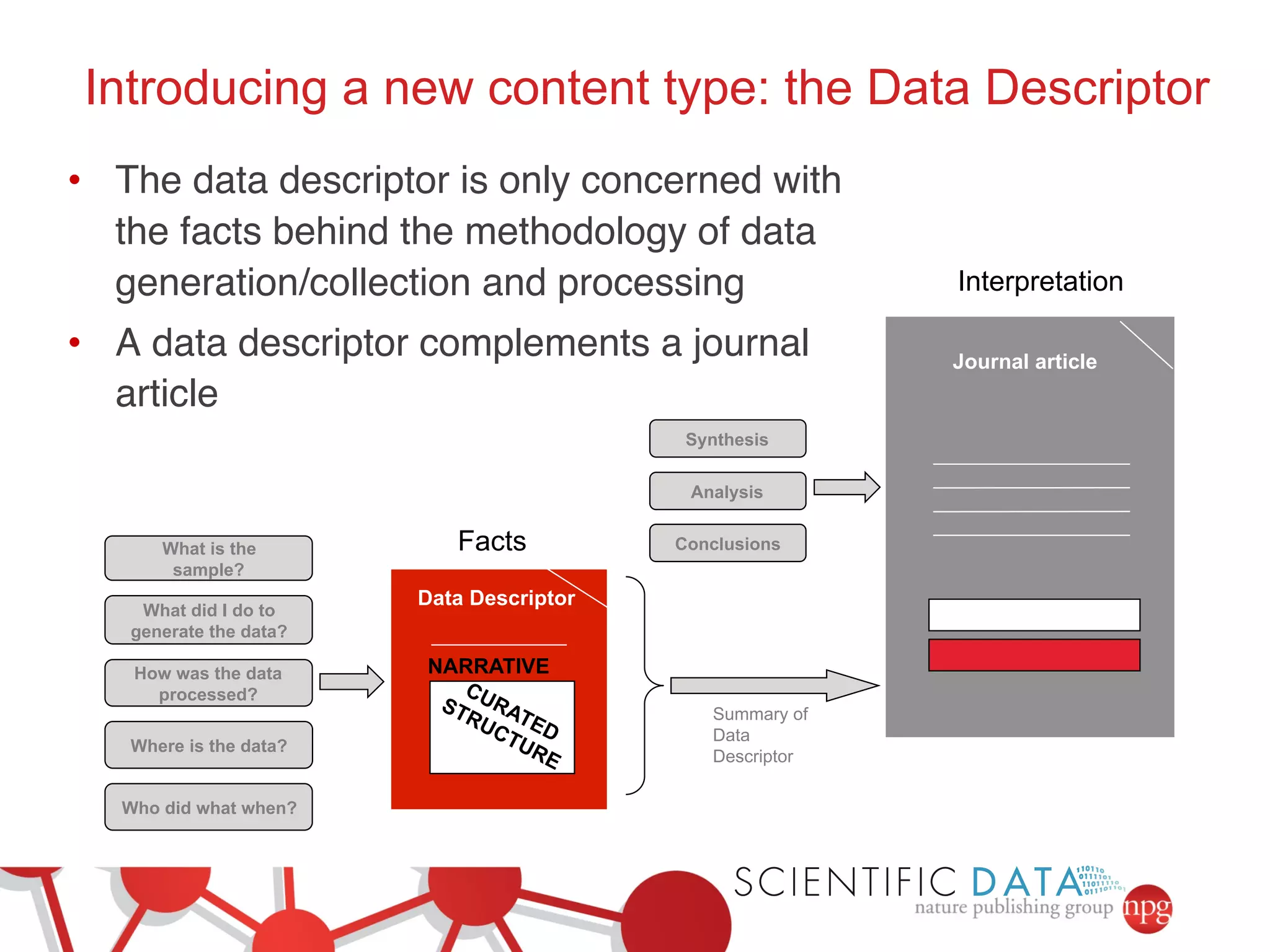
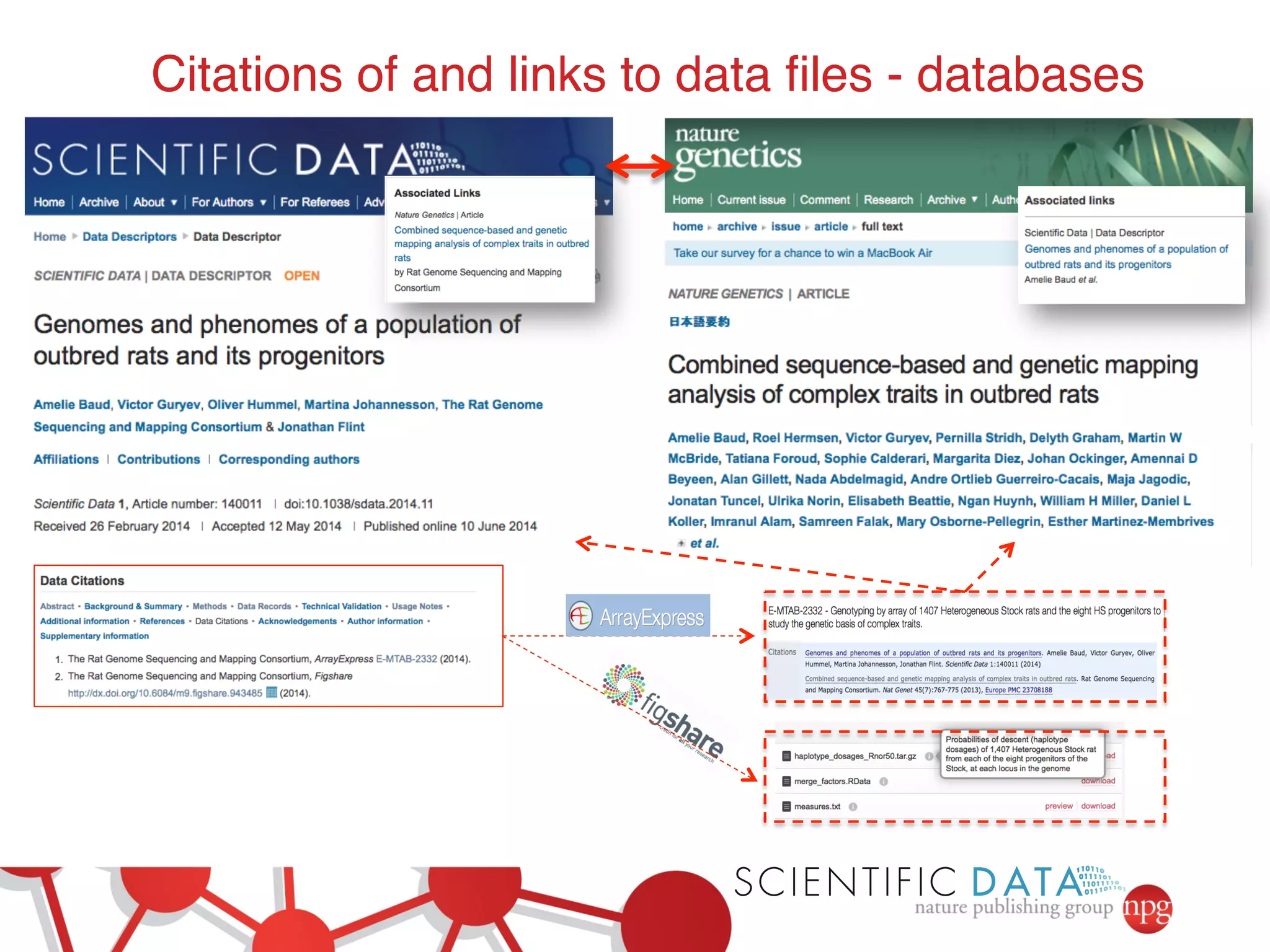
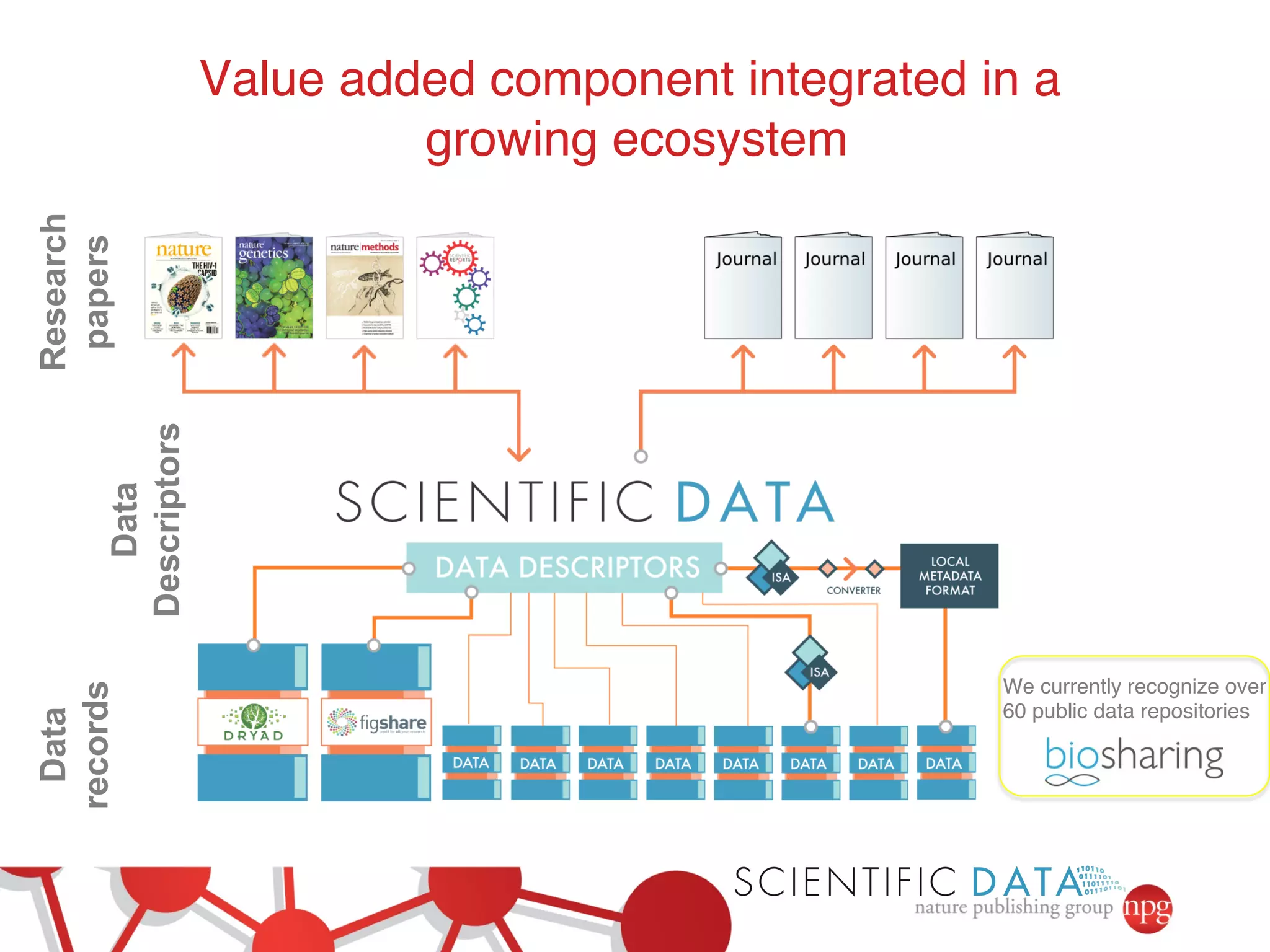
- The role of data journals in increasing credit/visibility for shared data and promoting standards/best practices.
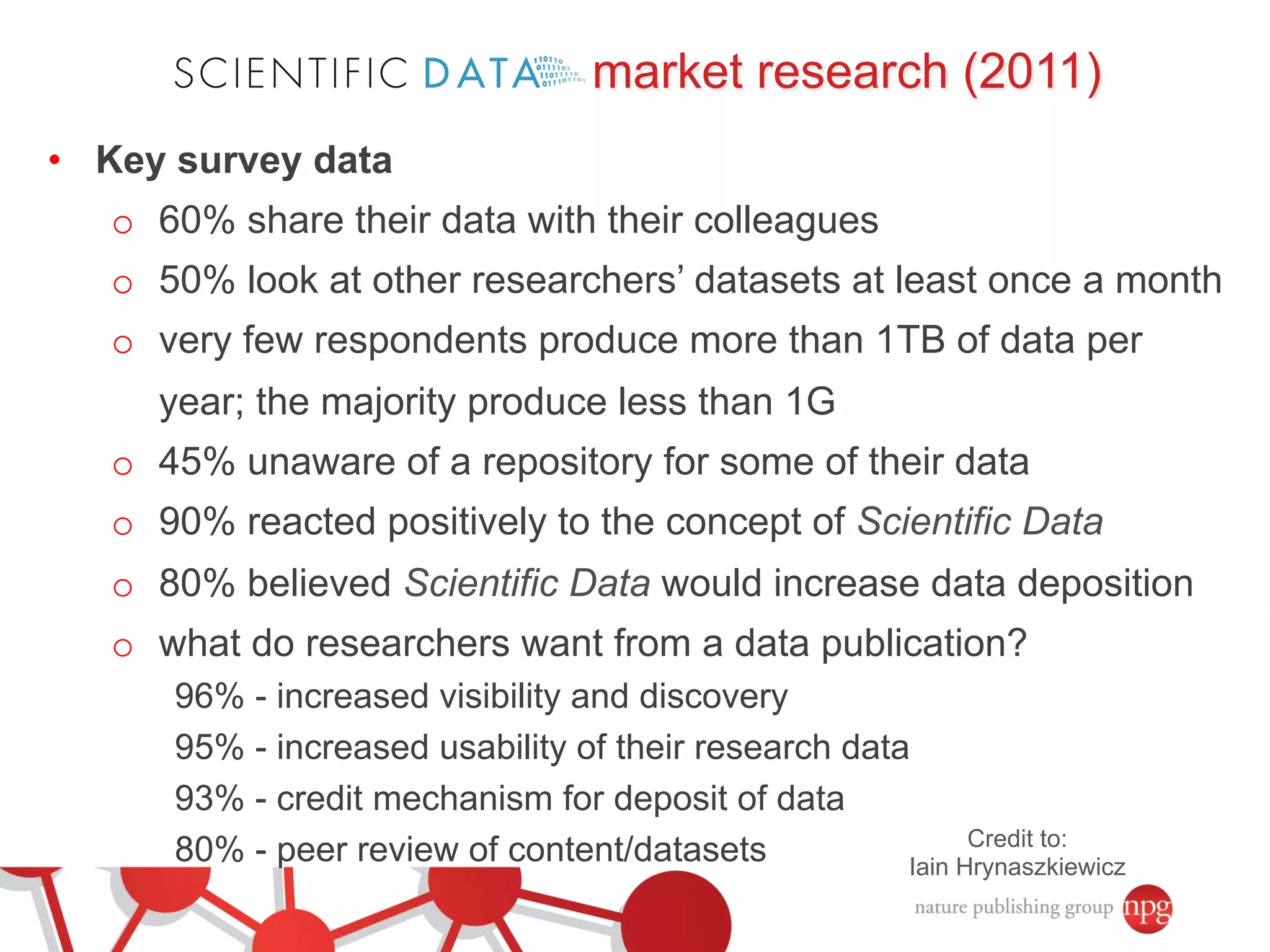
- Market research found researchers want increased visibility, usability, and credit for sharing their data.