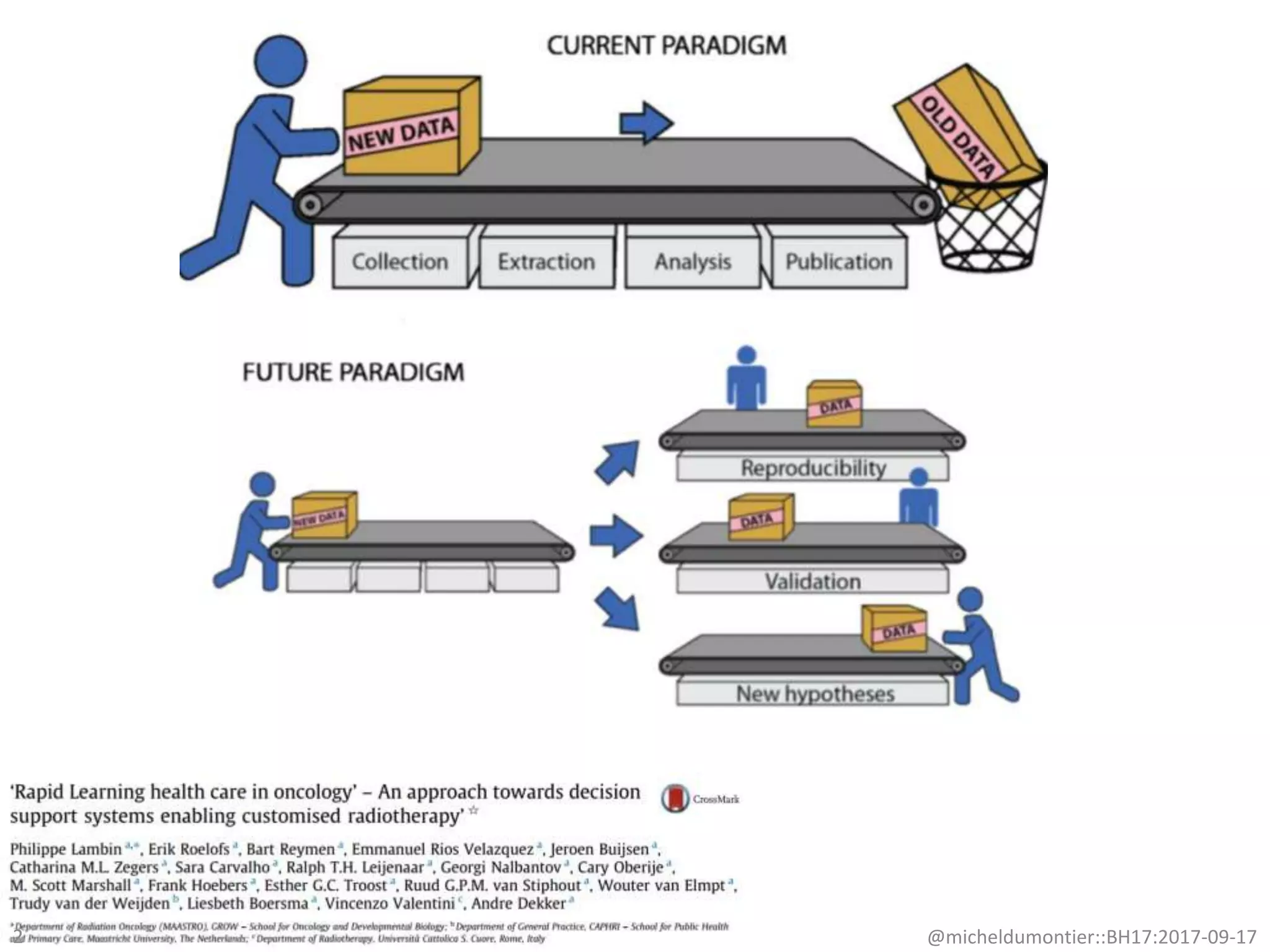
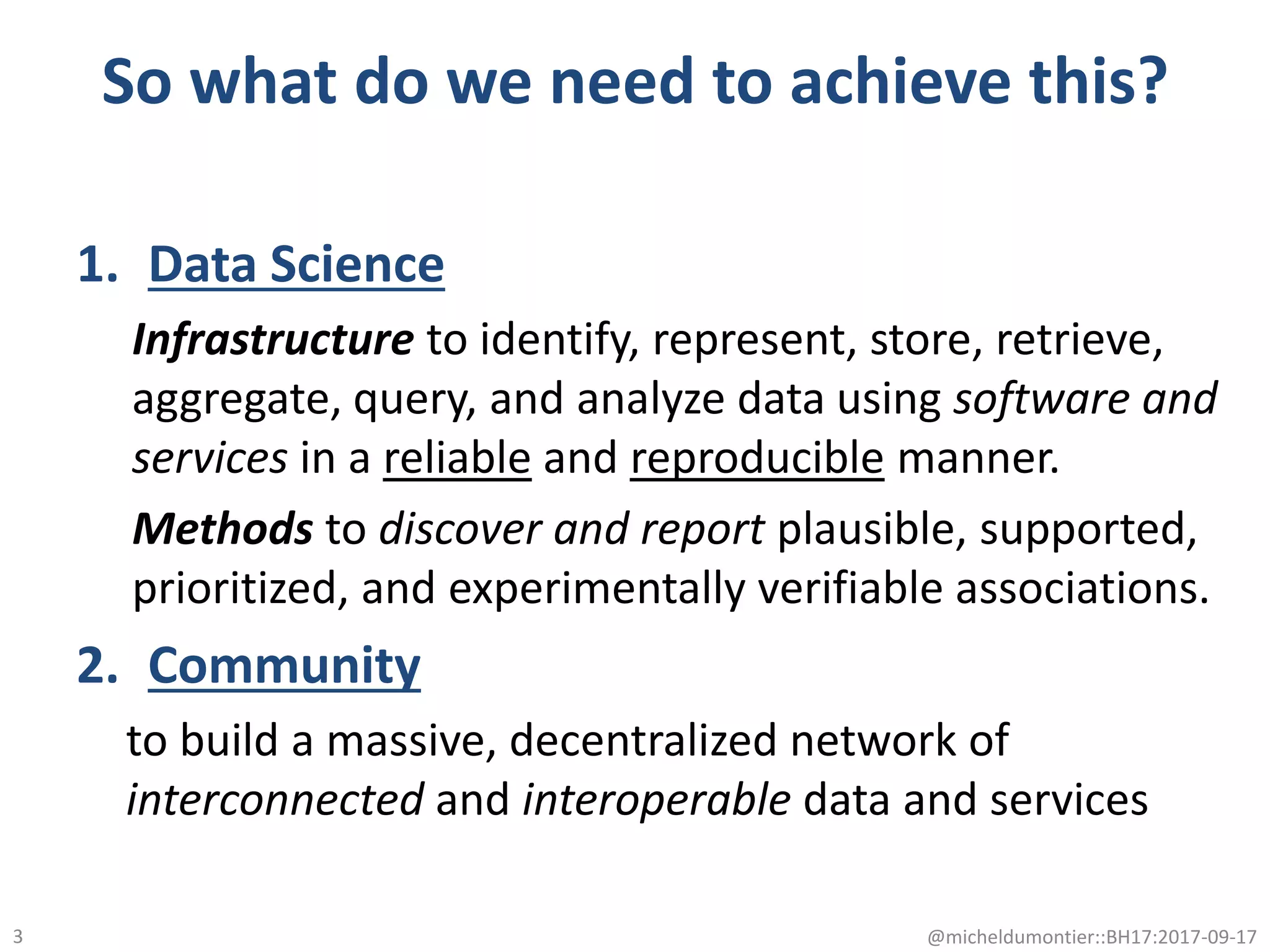
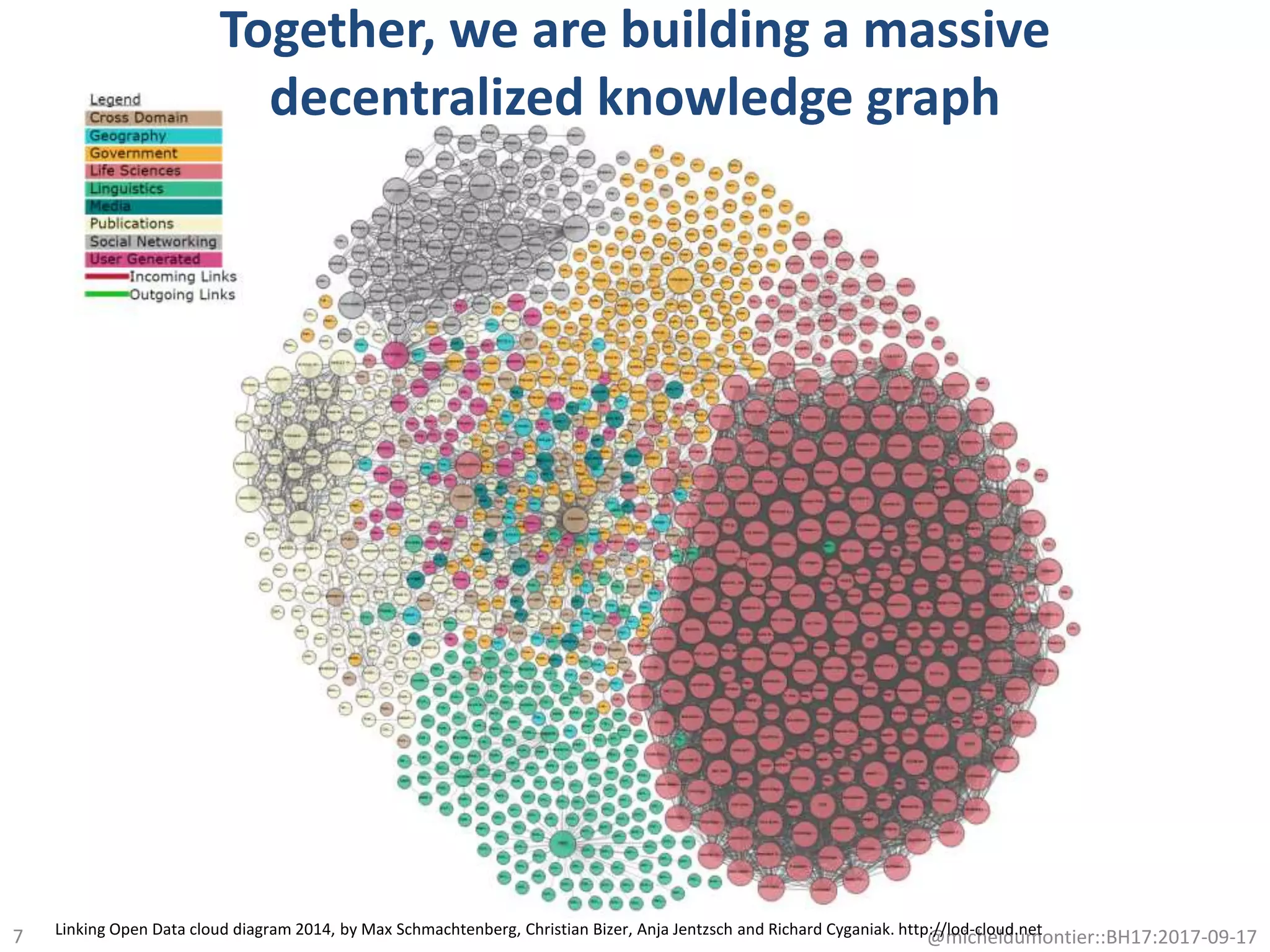
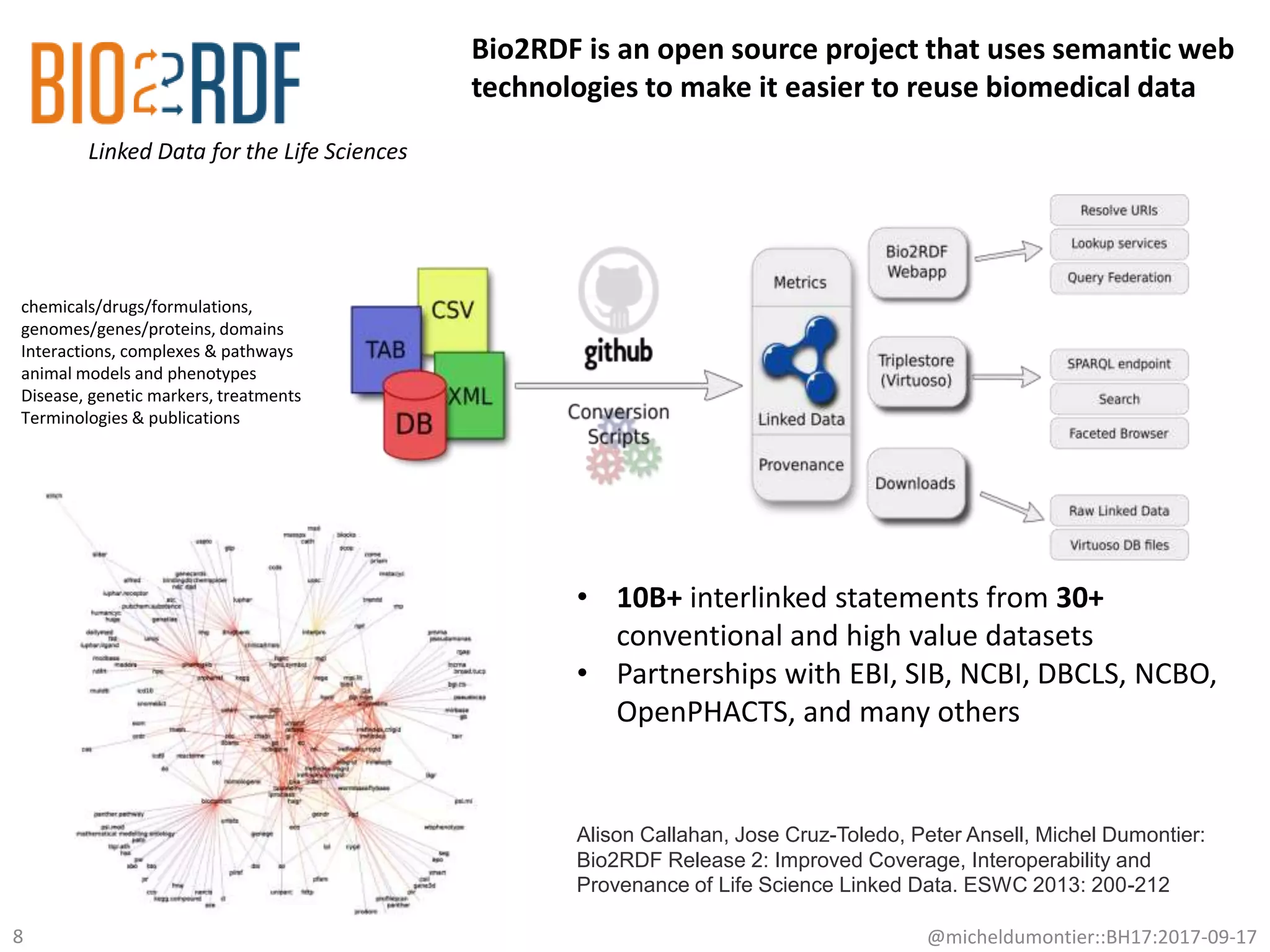
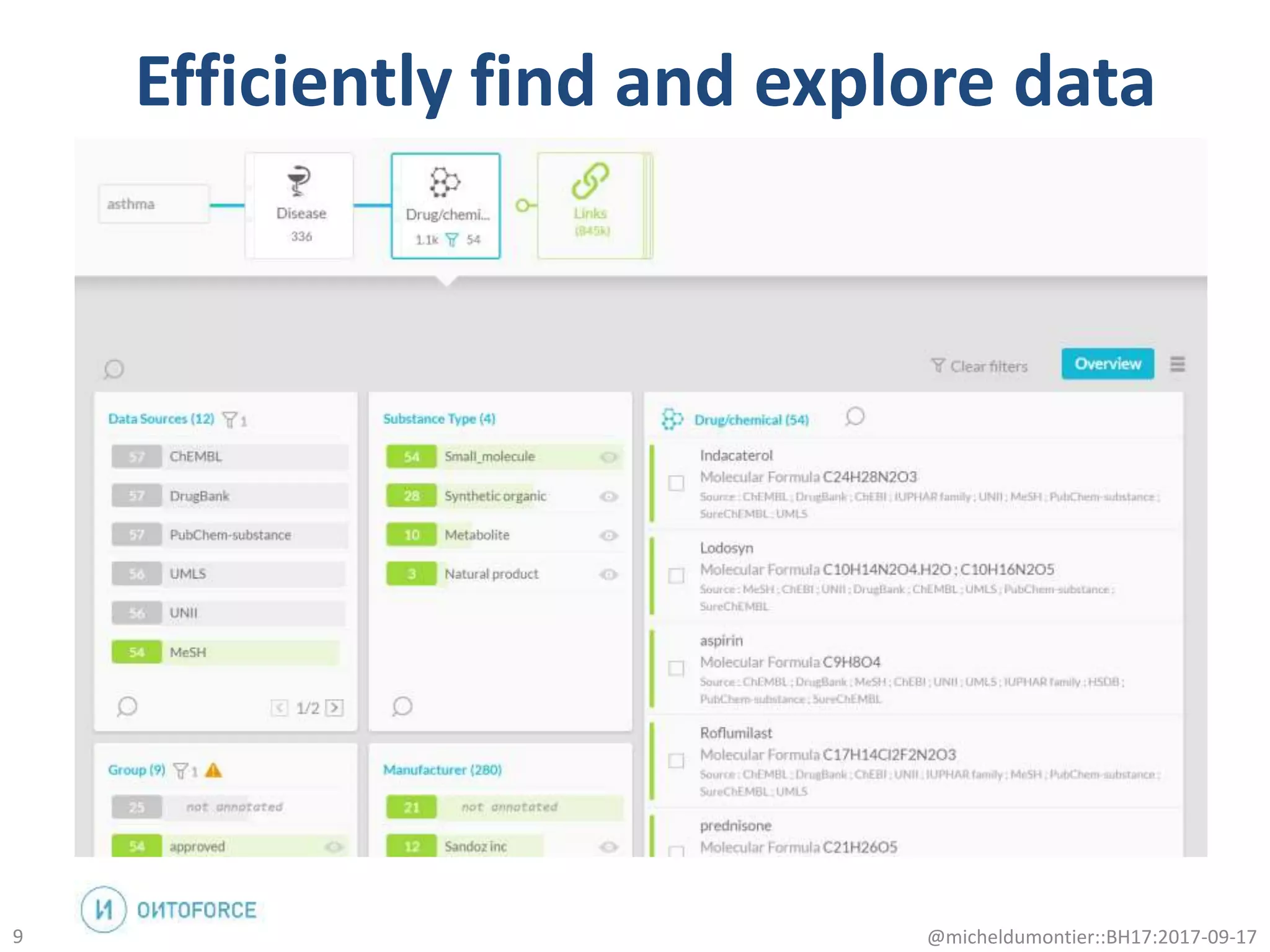
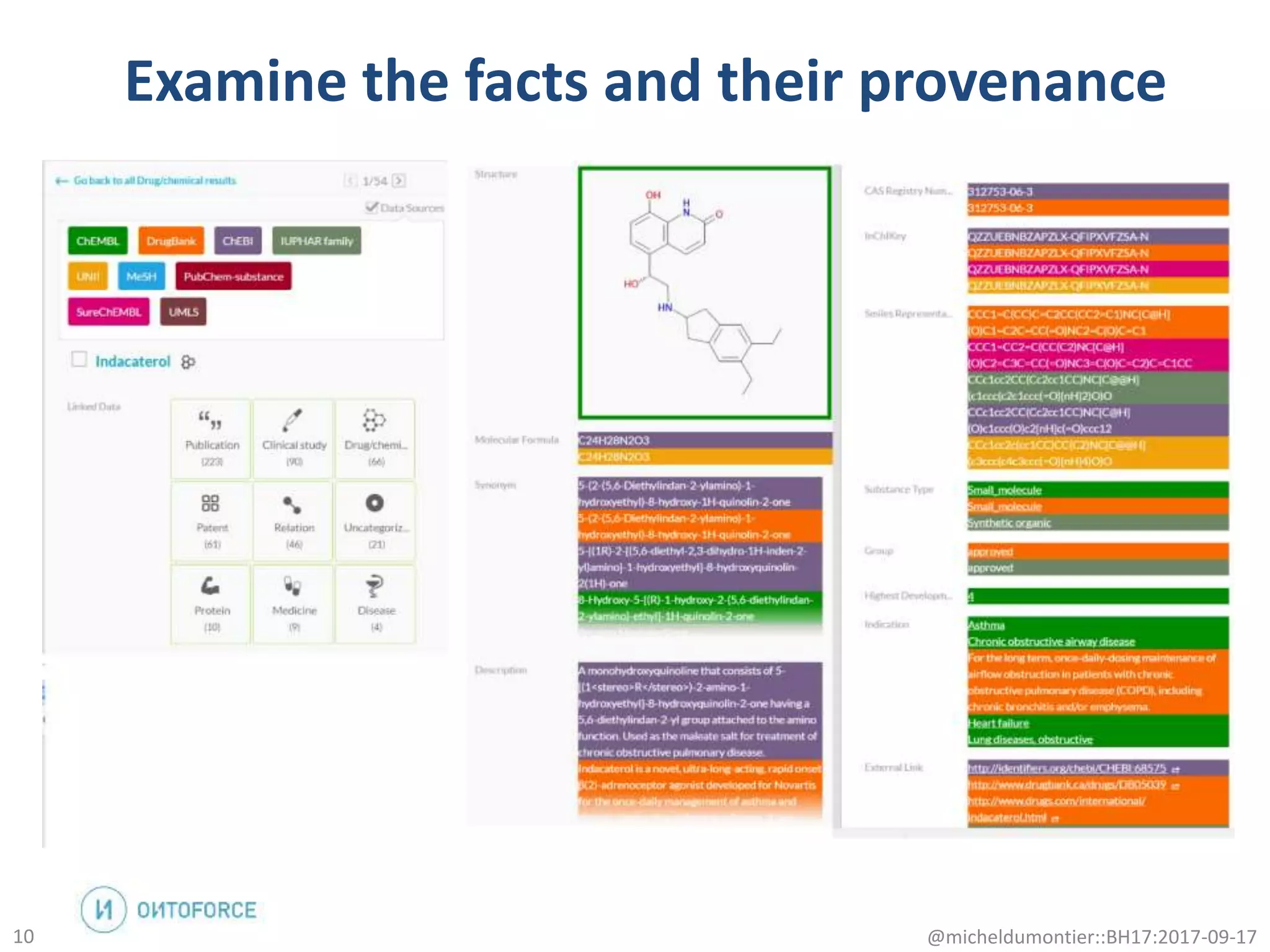
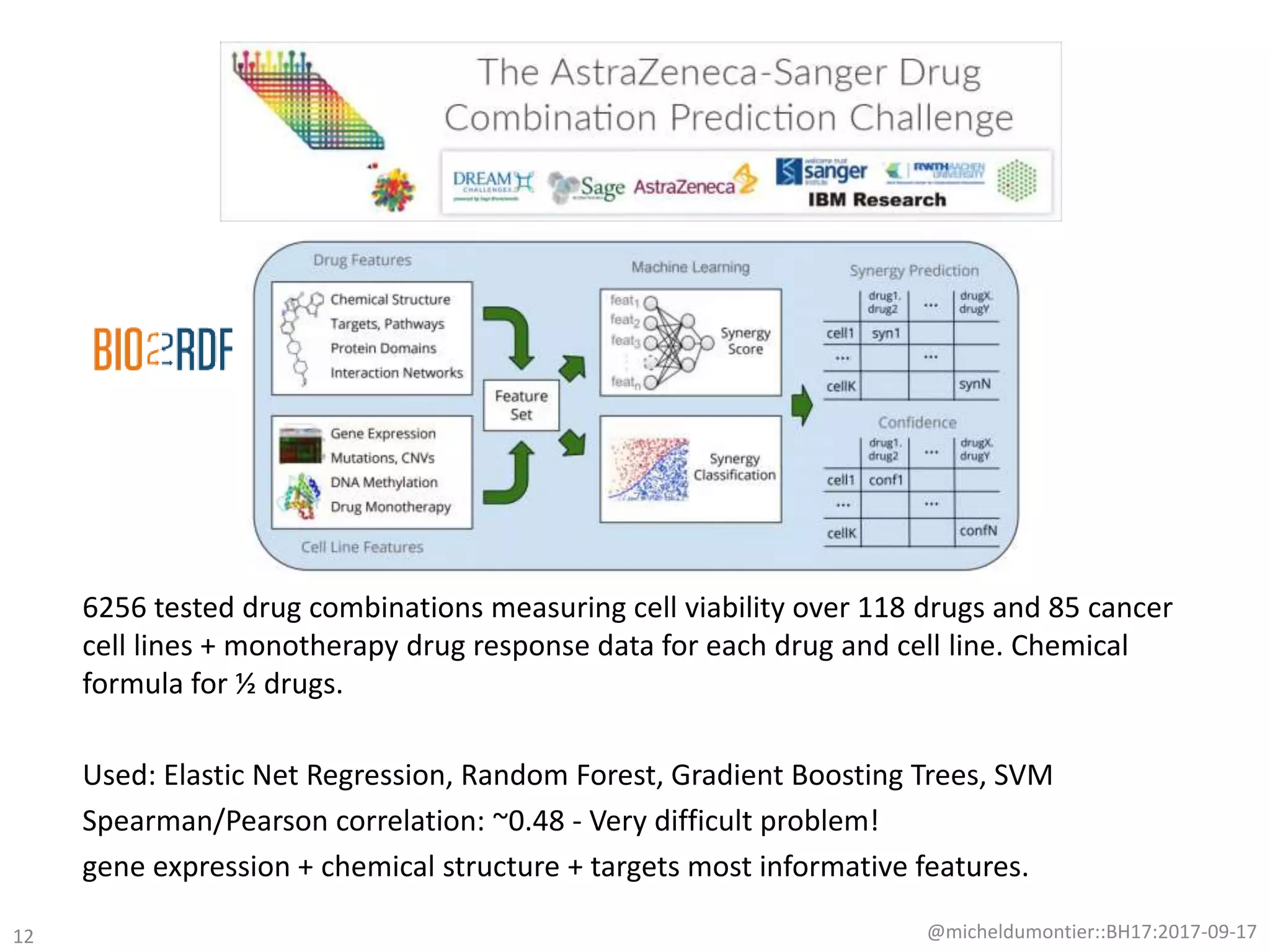
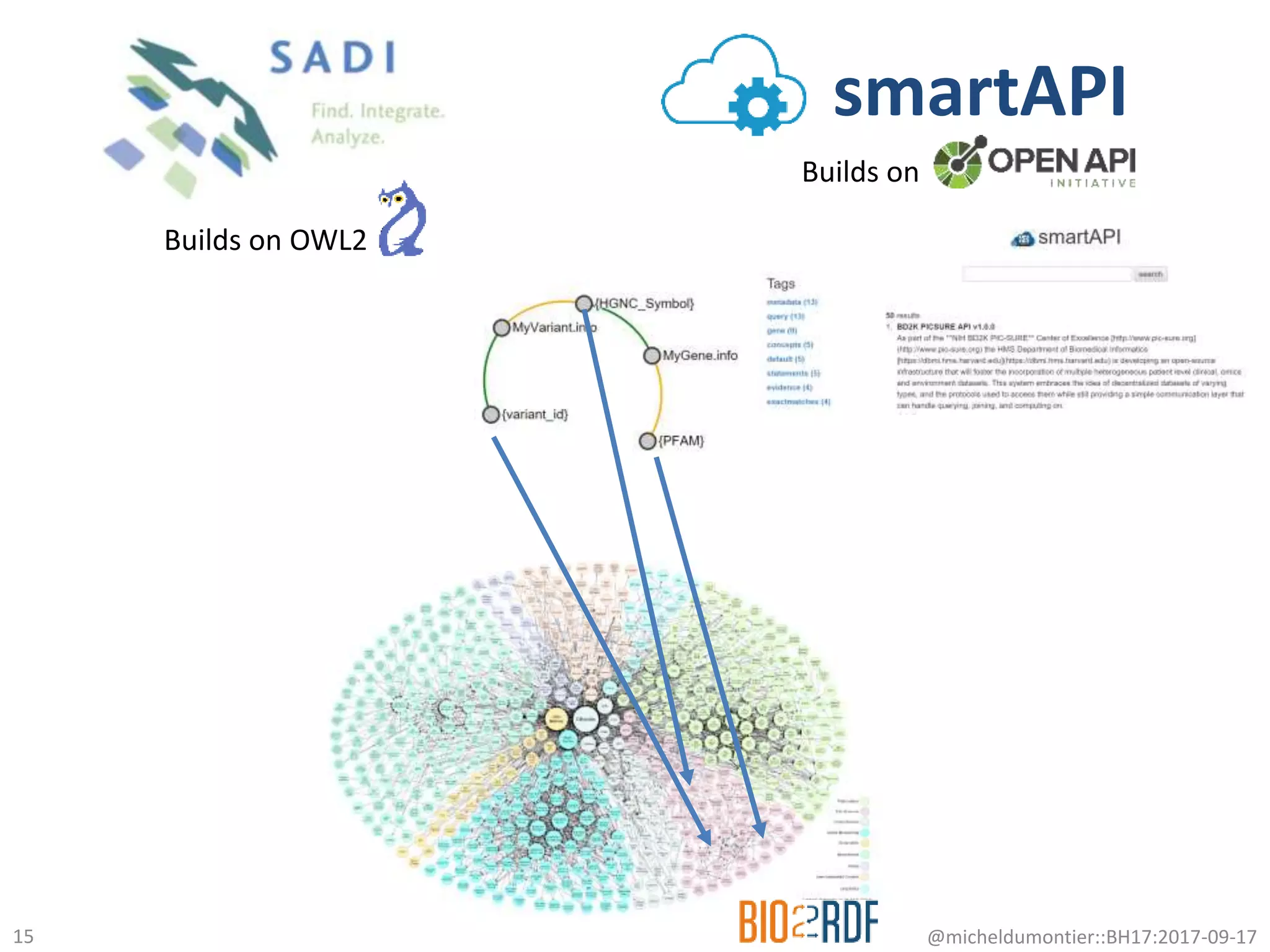
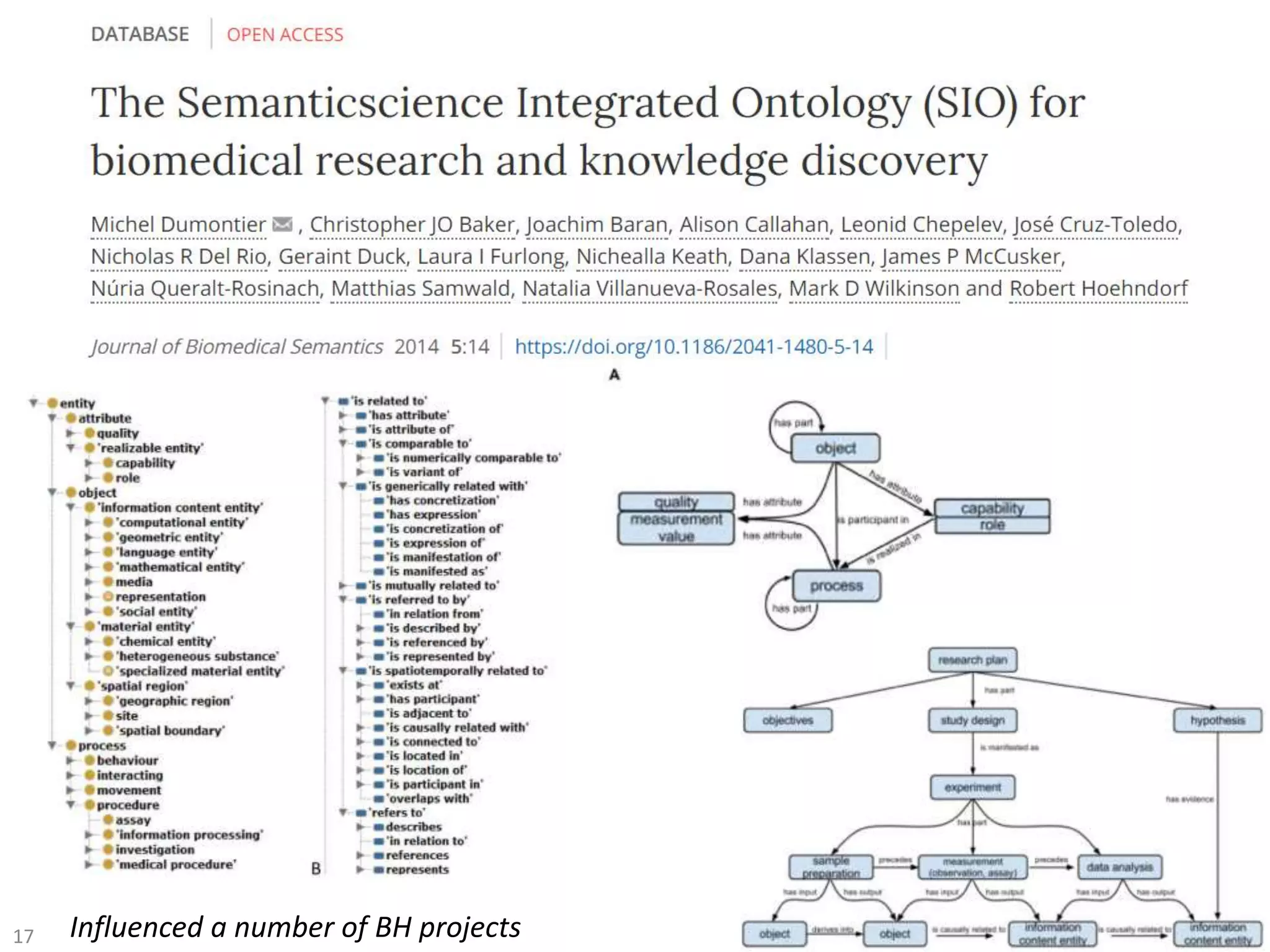
The document discusses the advancement of biomedical knowledge through the implementation of FAIR principles for data sharing and reuse. It emphasizes the need for a robust data science infrastructure, community collaboration, and high-quality metadata to facilitate reliable access to biomedical data. The document also outlines ongoing projects and goals related to improving the interoperability, discoverability, and overall efficiency of biomedical data utilization.