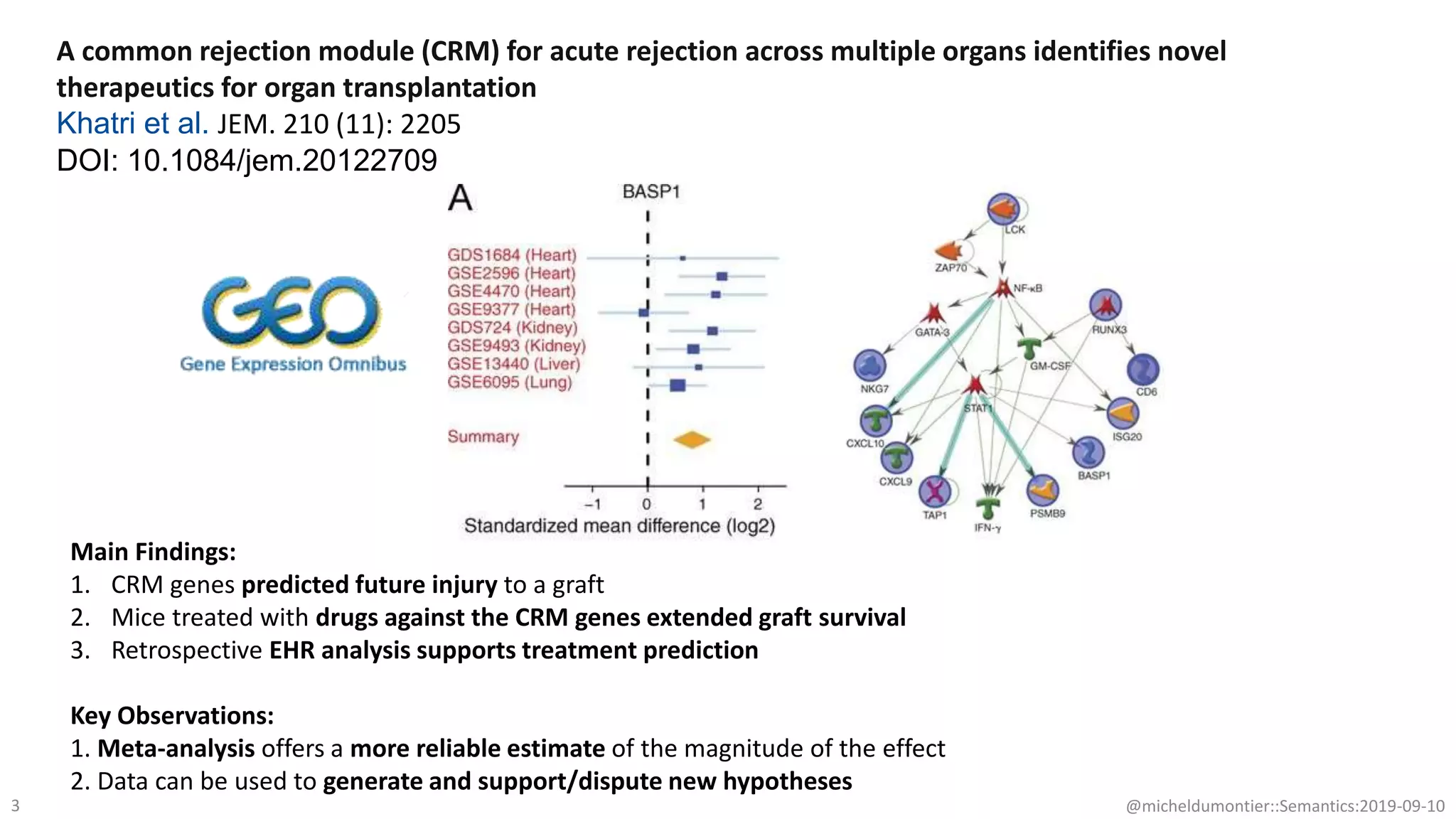
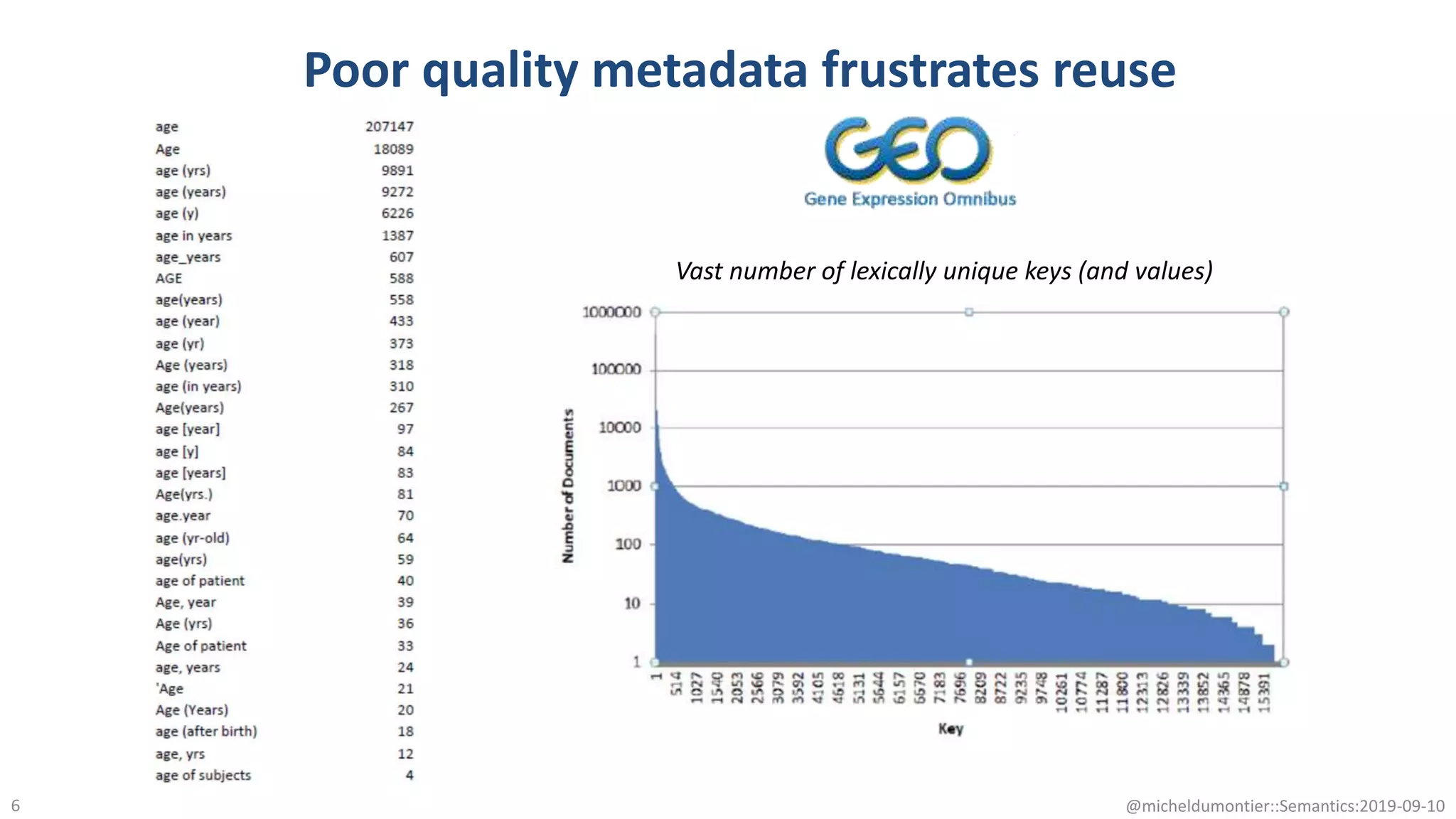
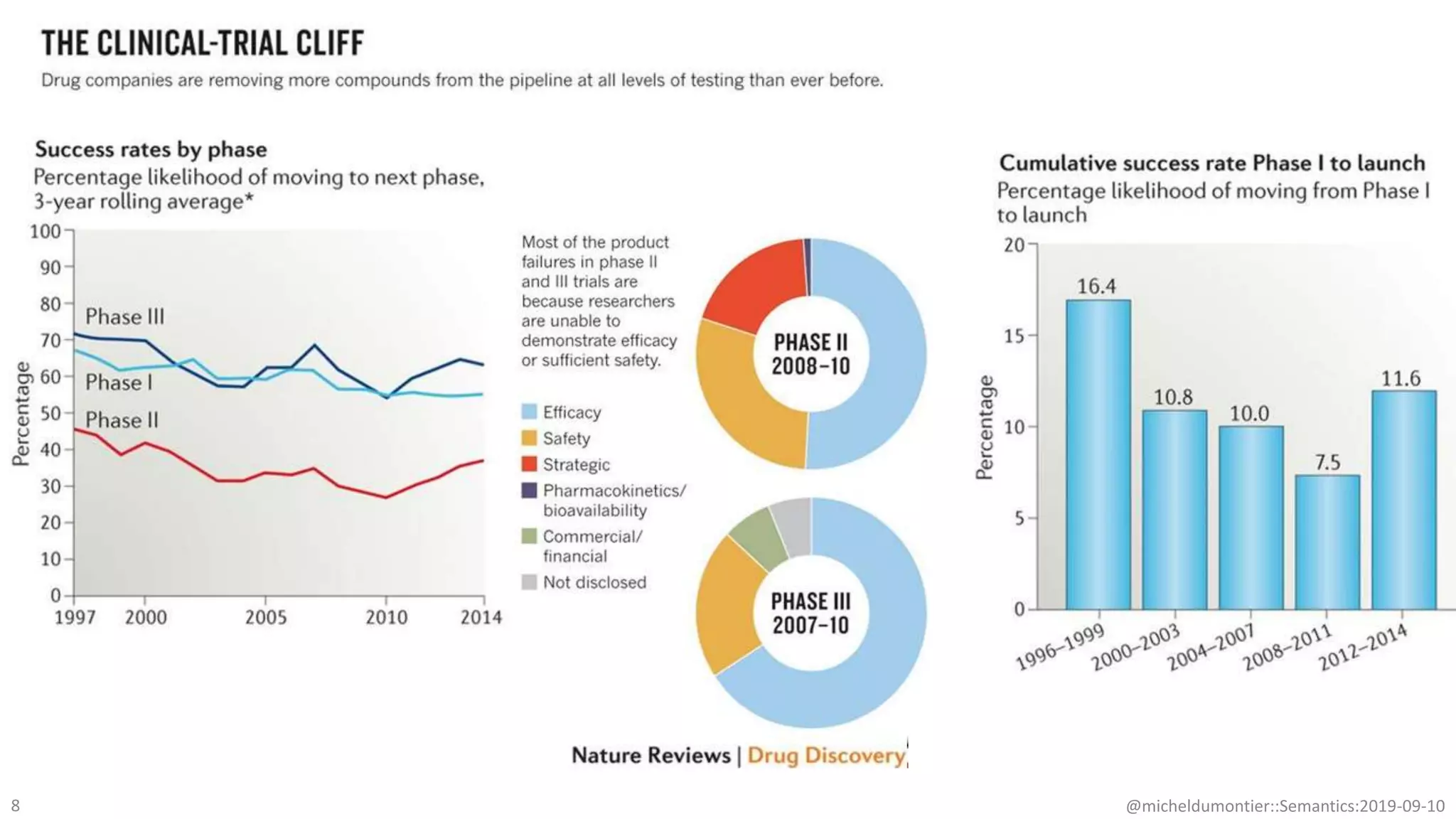
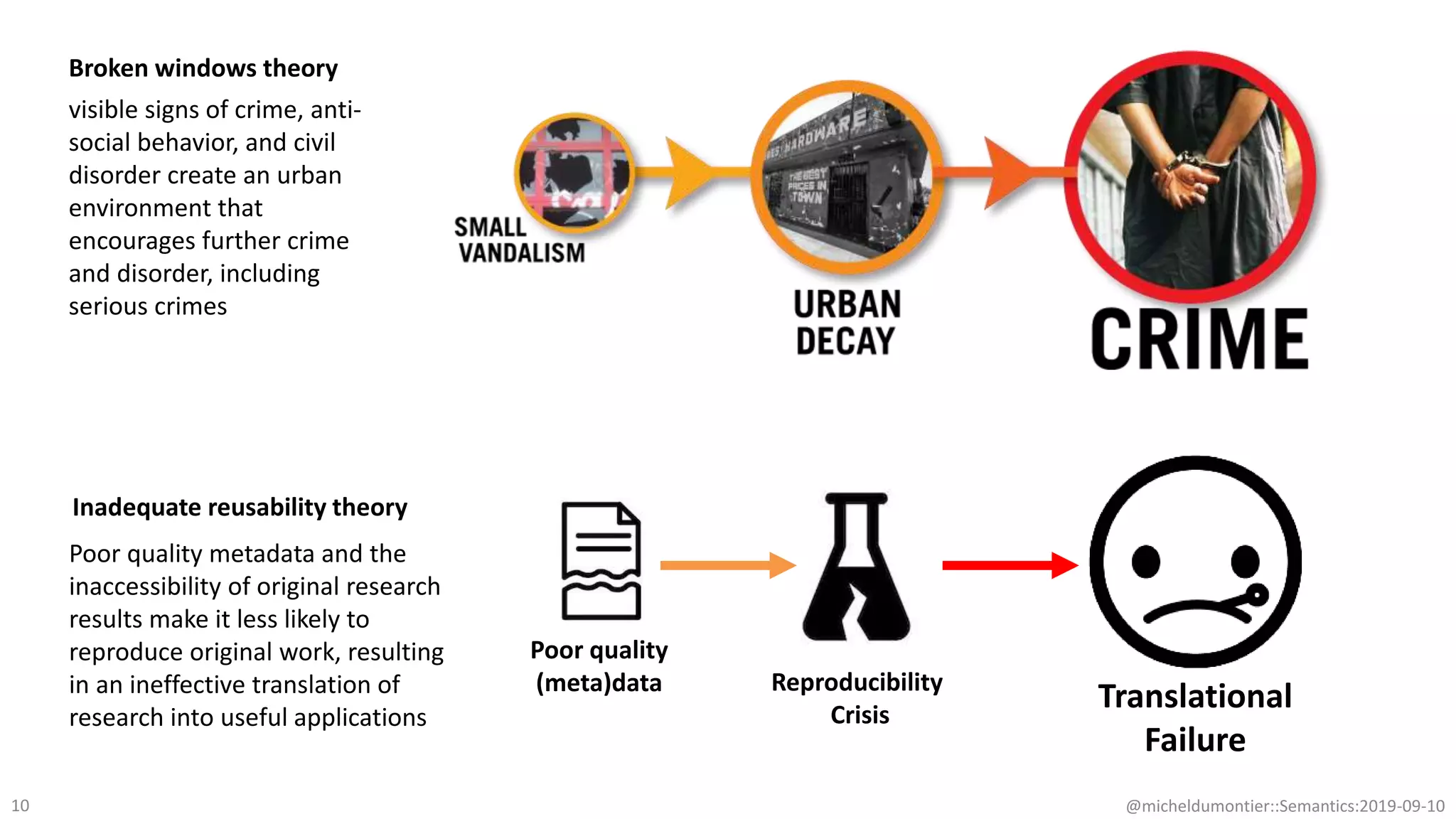
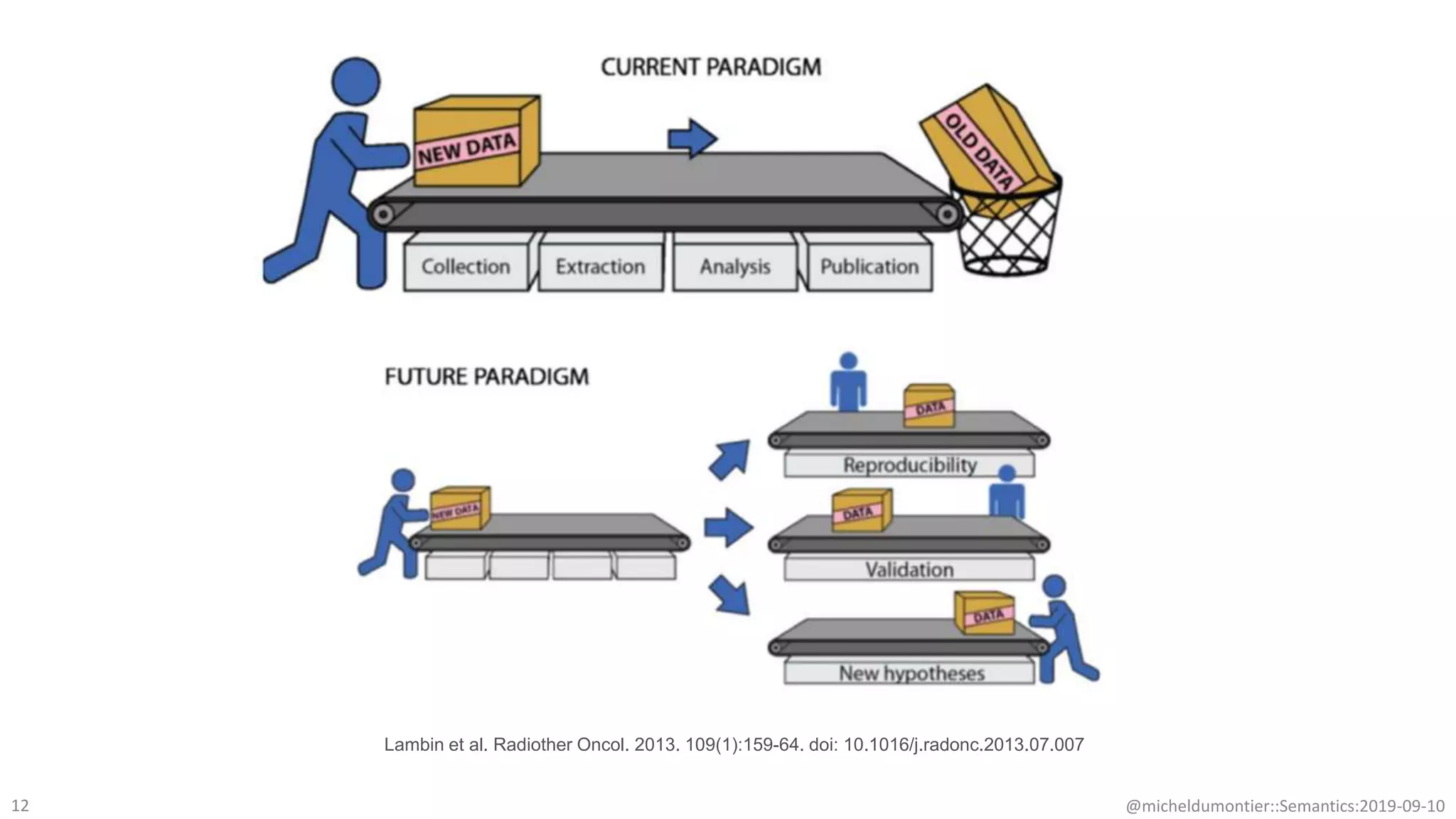
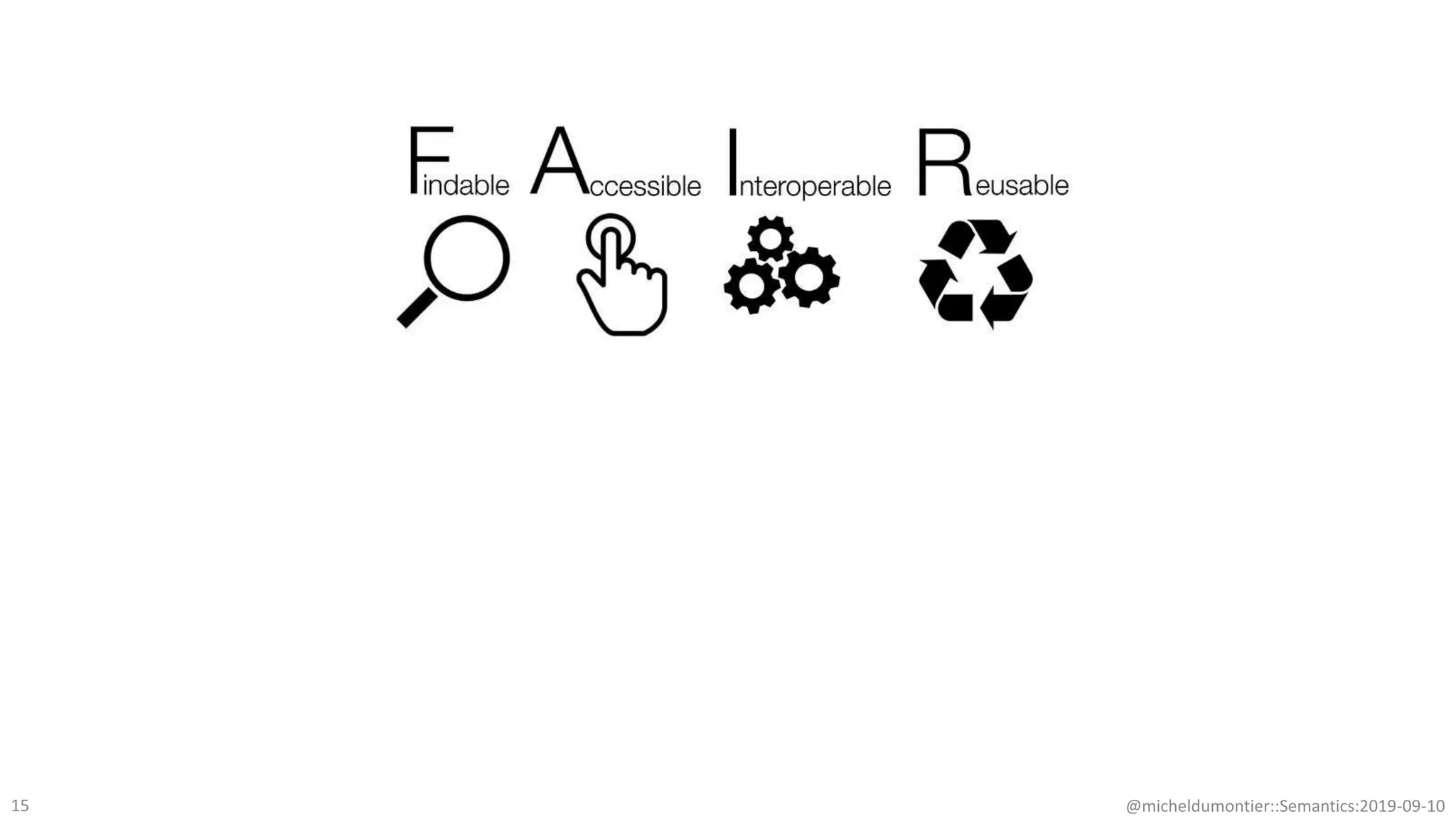
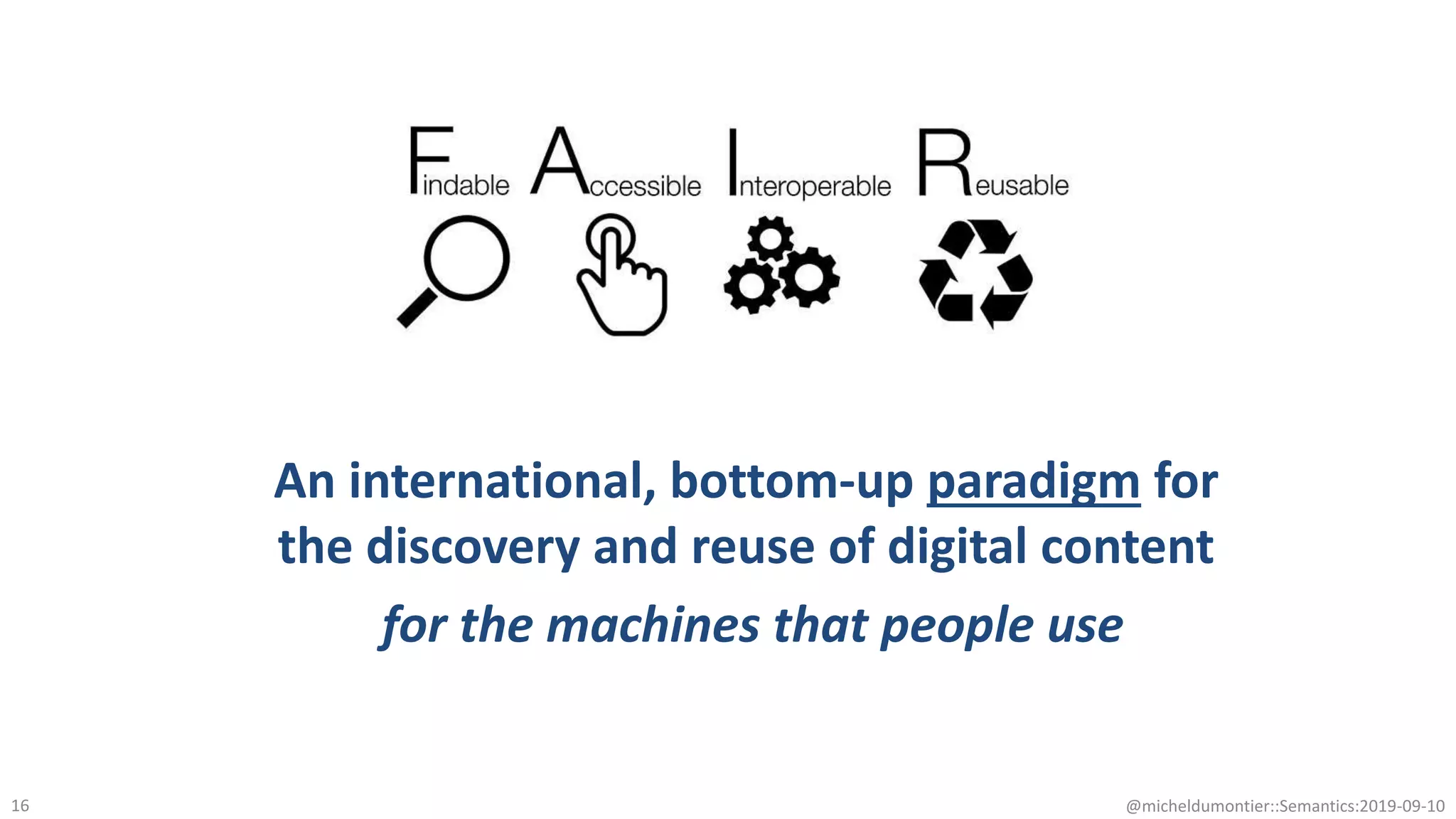
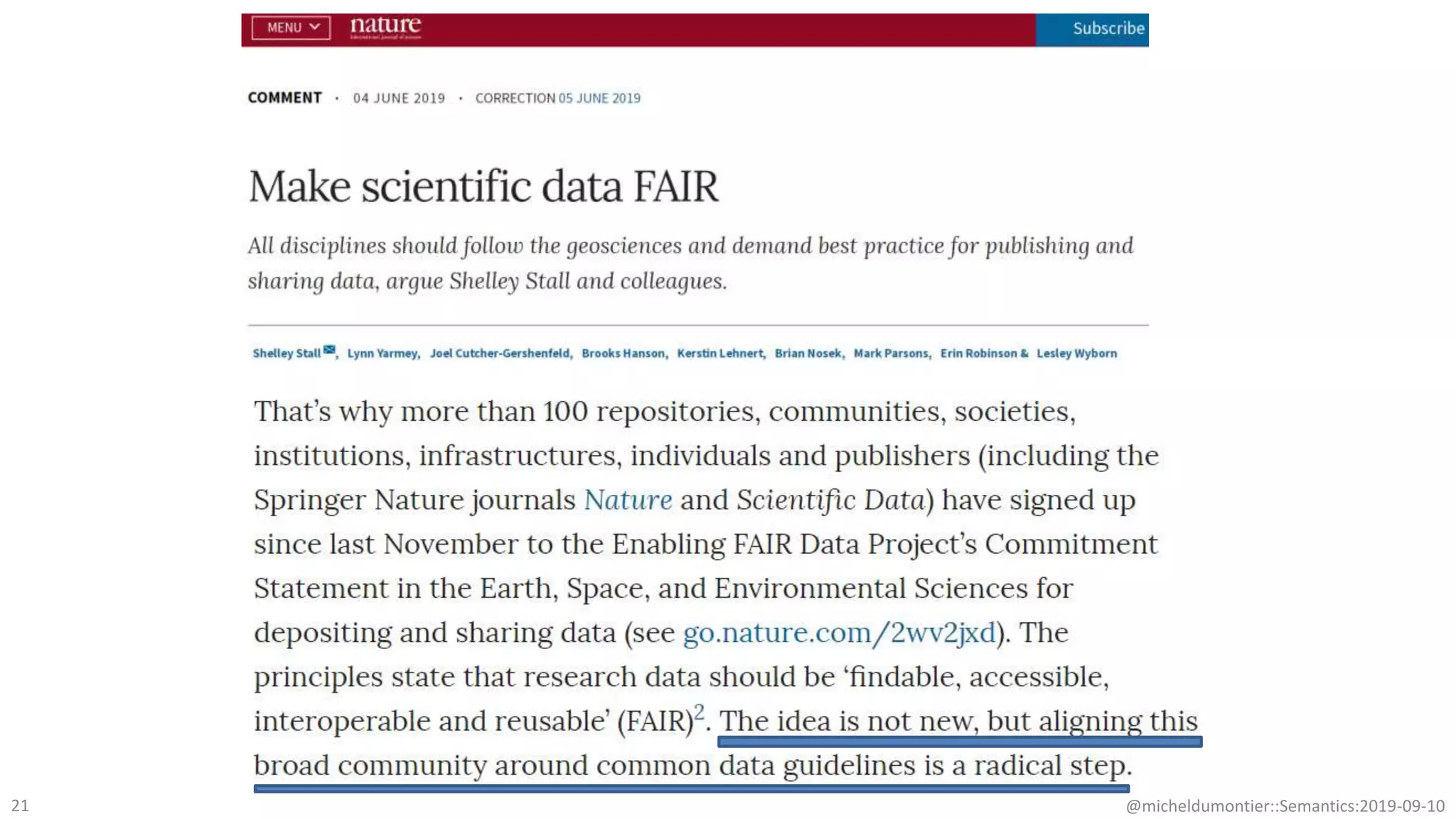
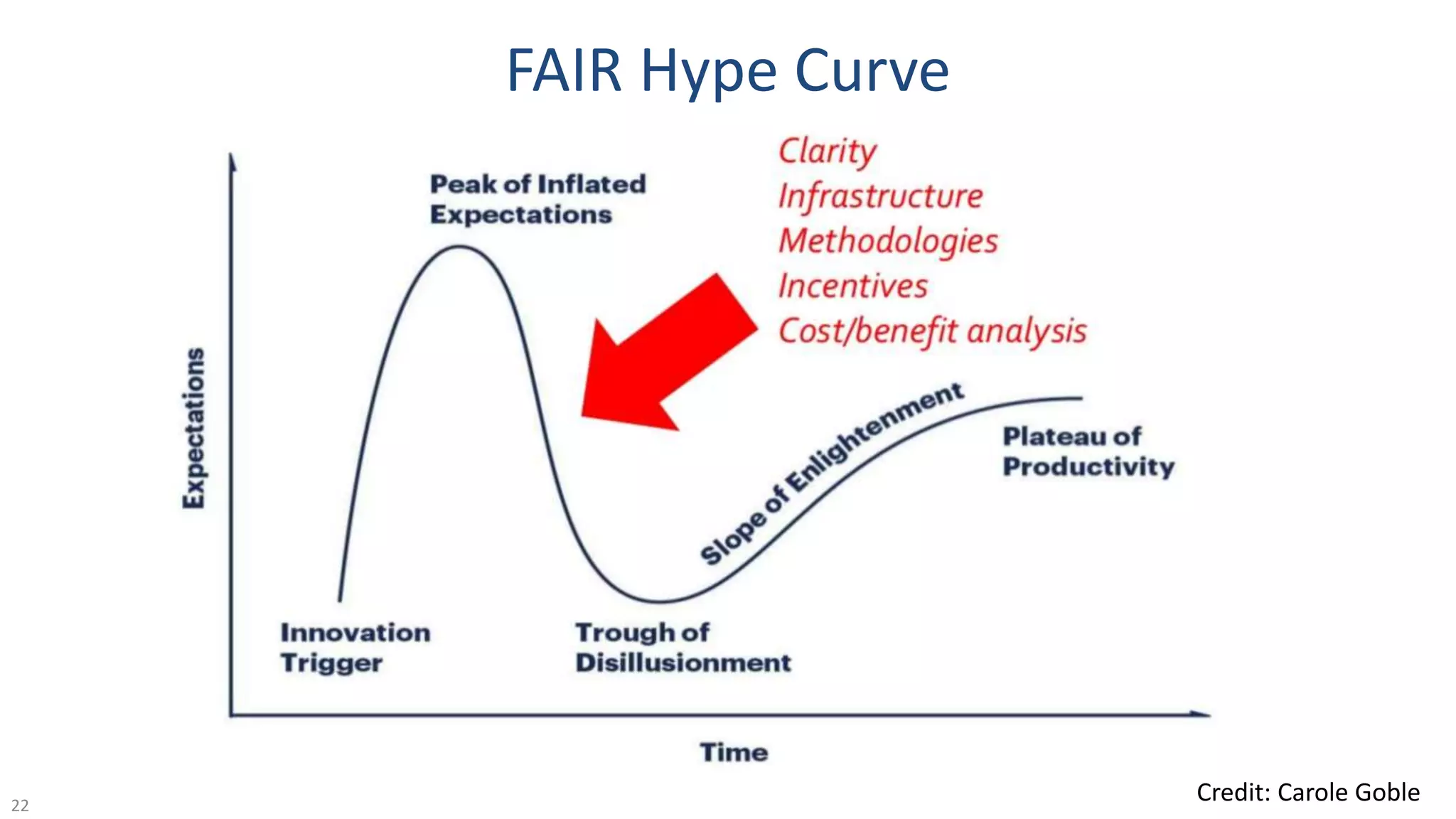
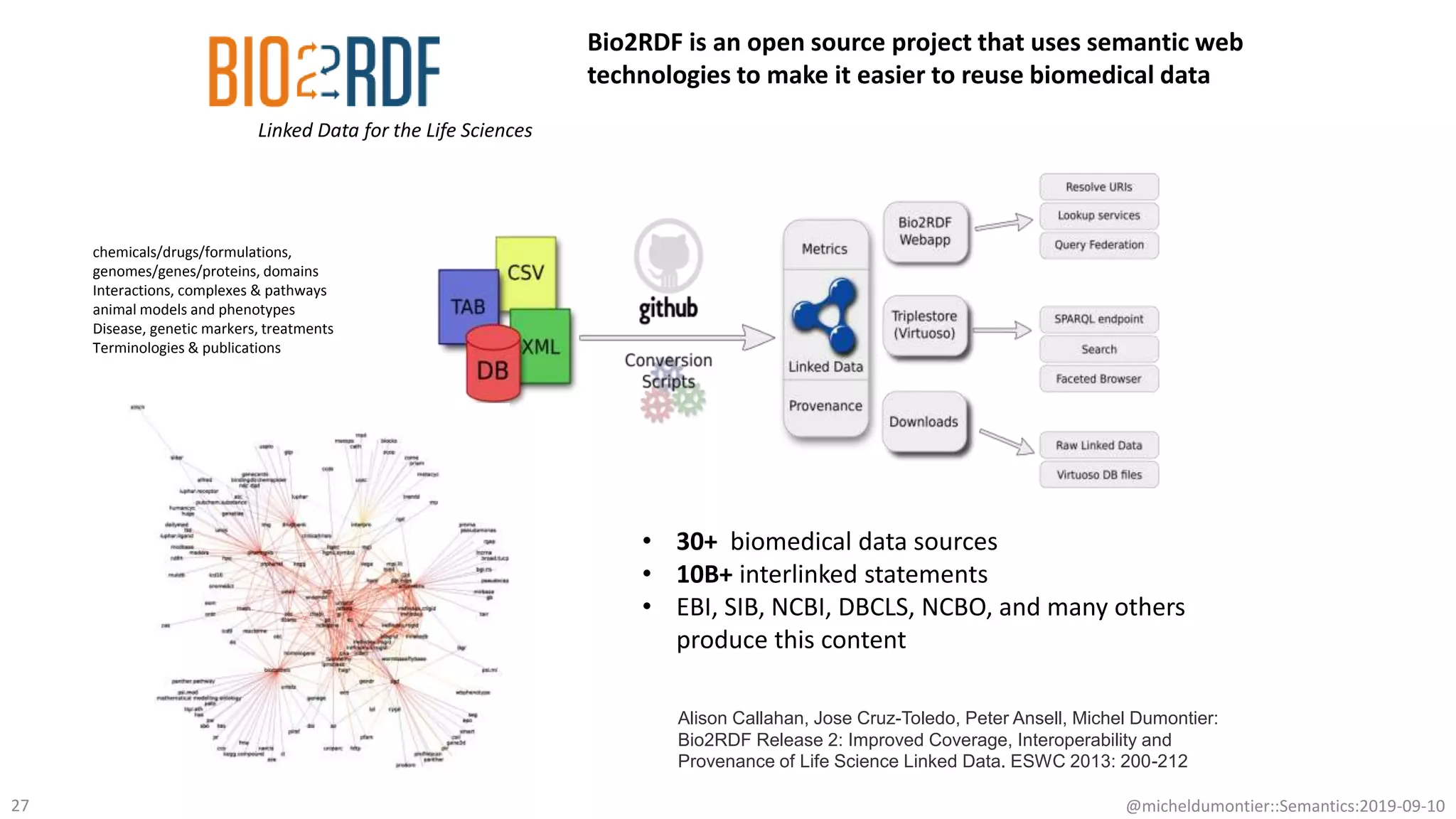
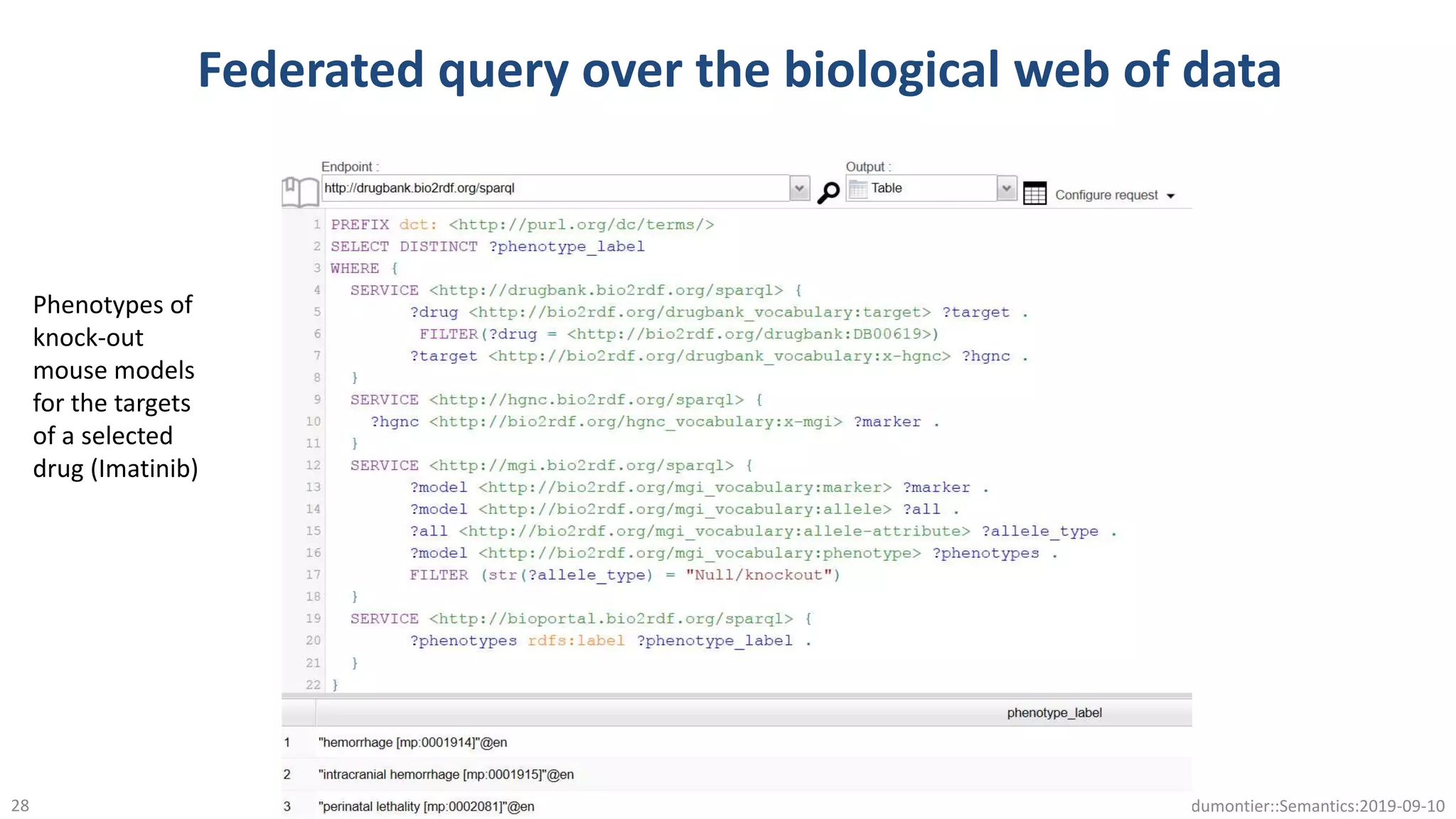
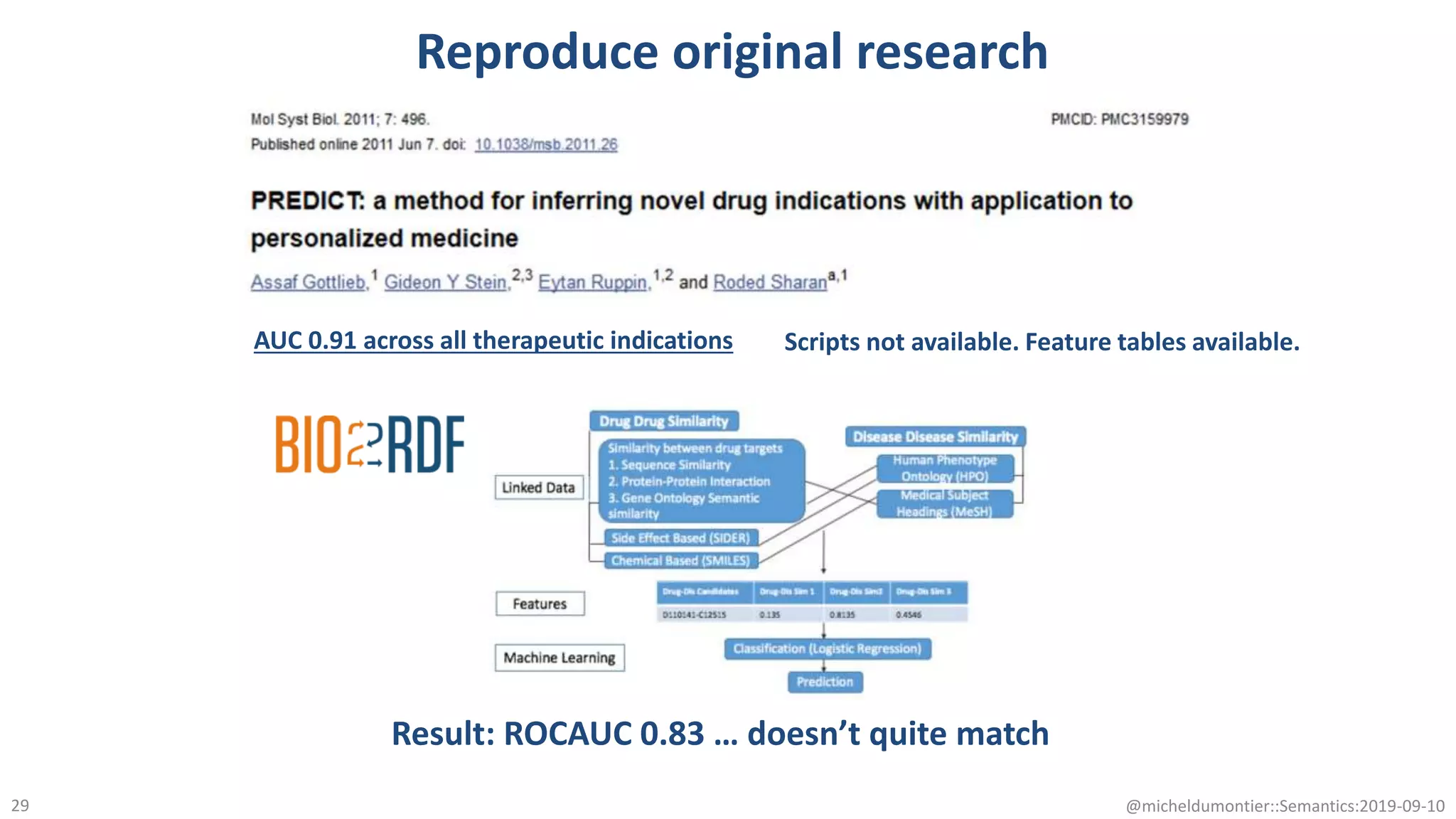
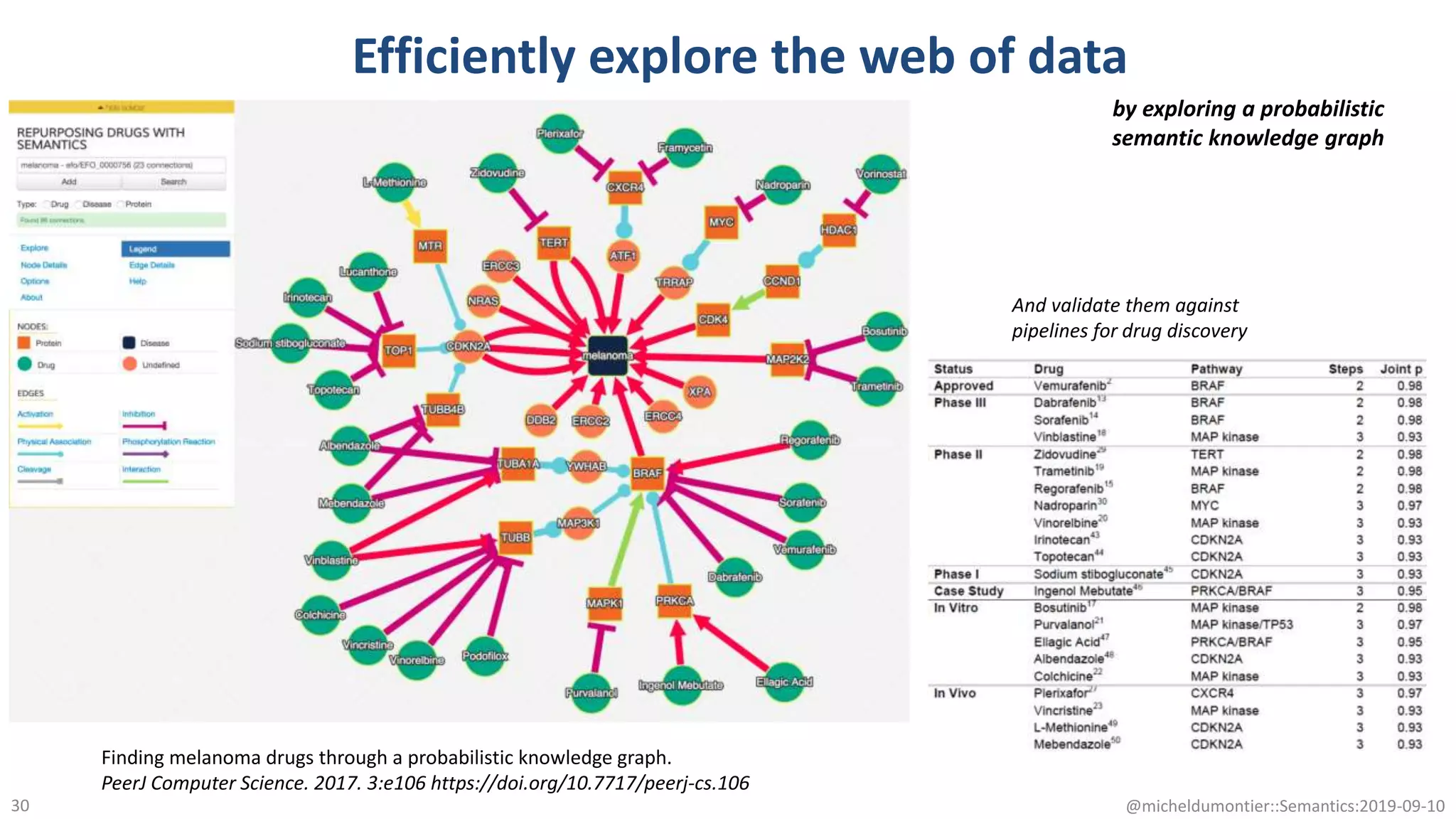
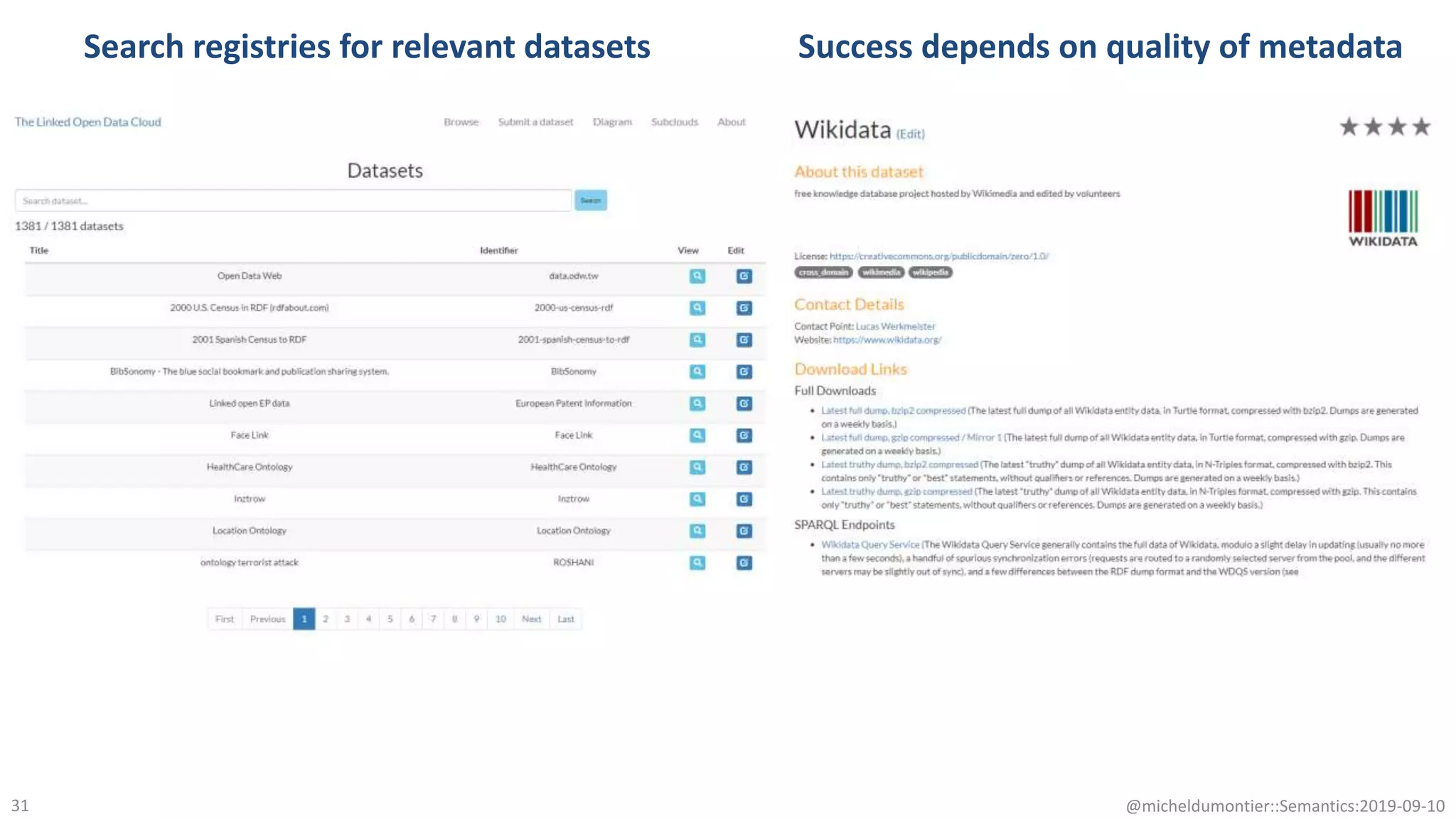
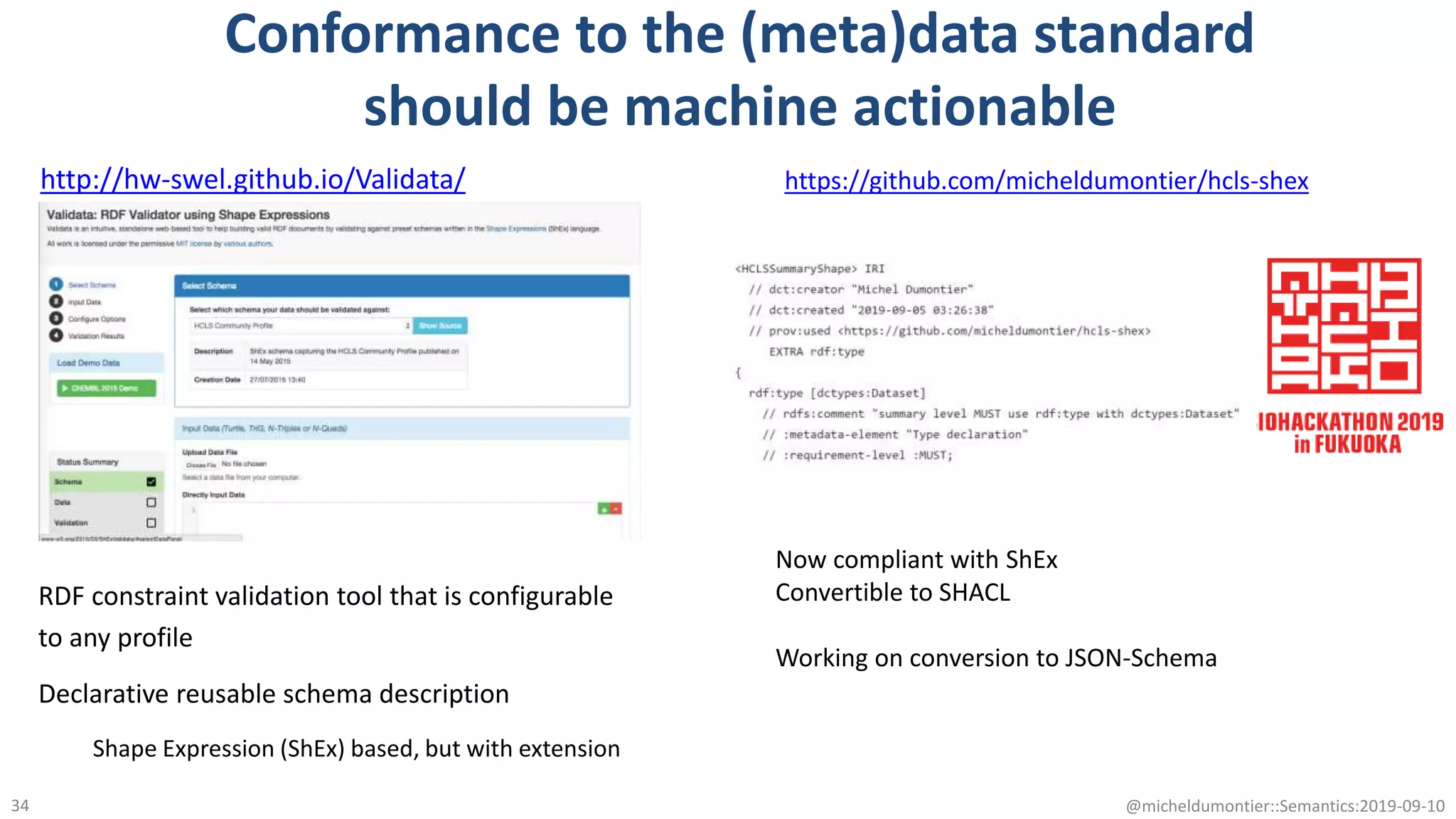
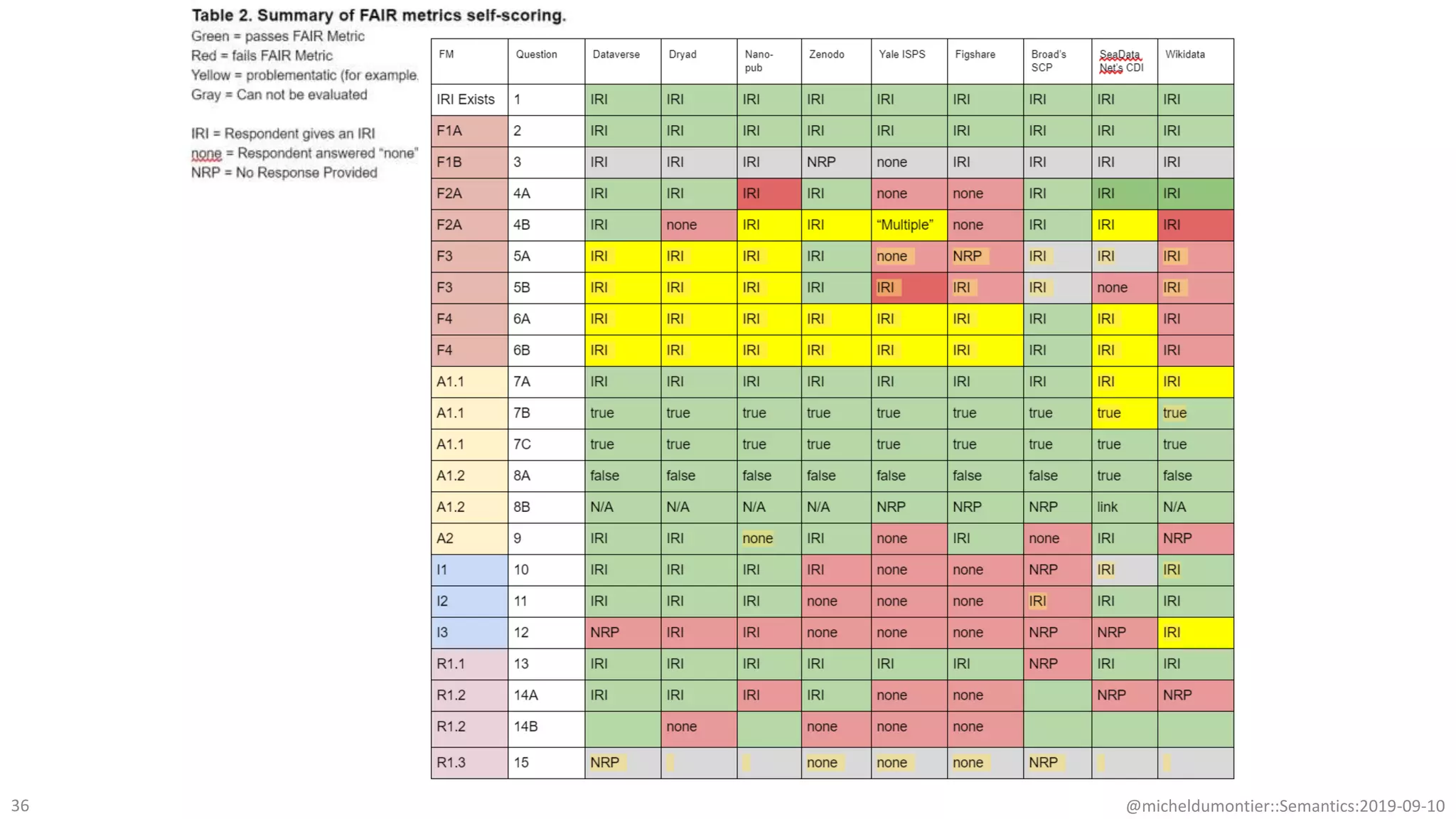
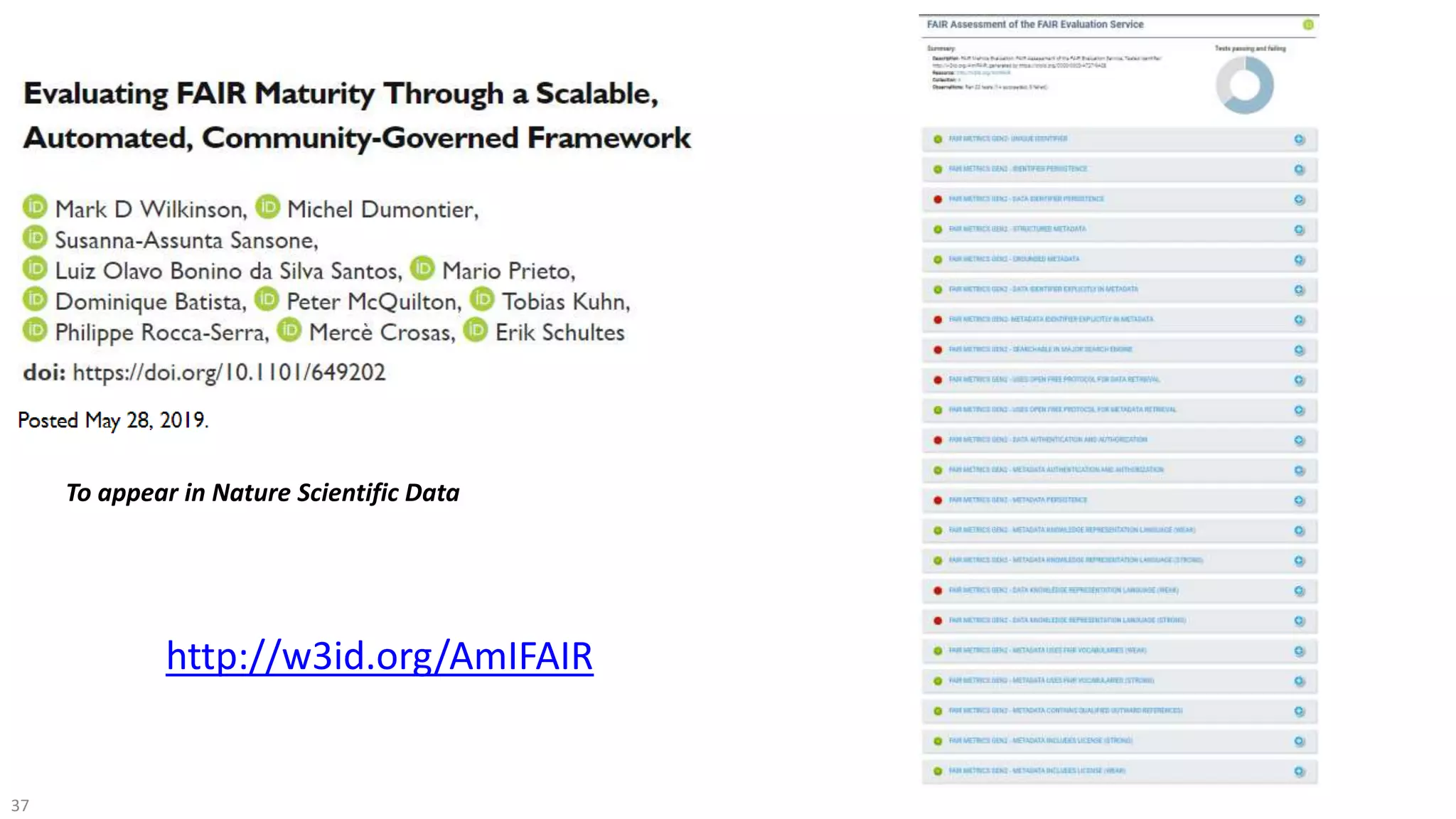
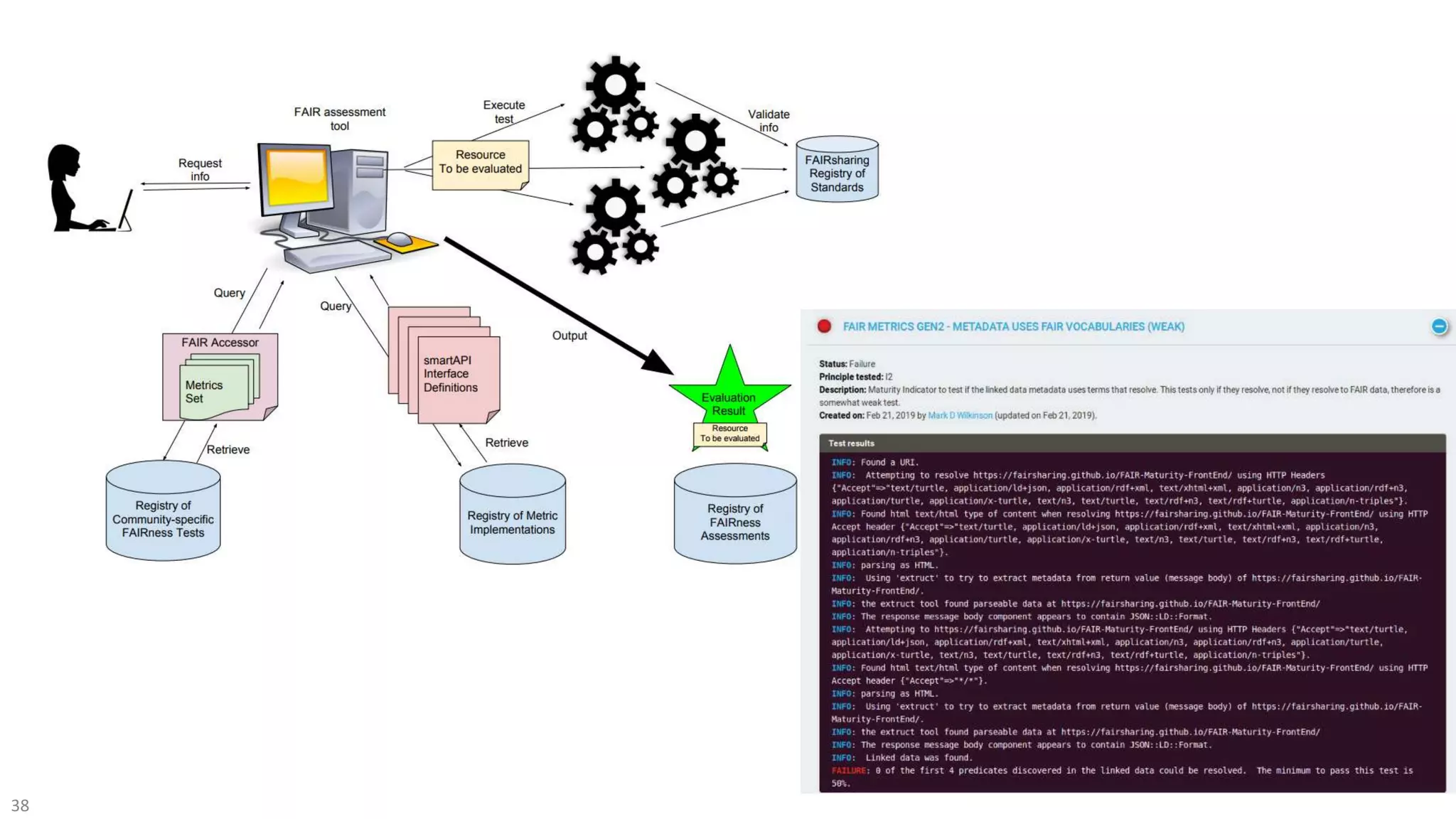
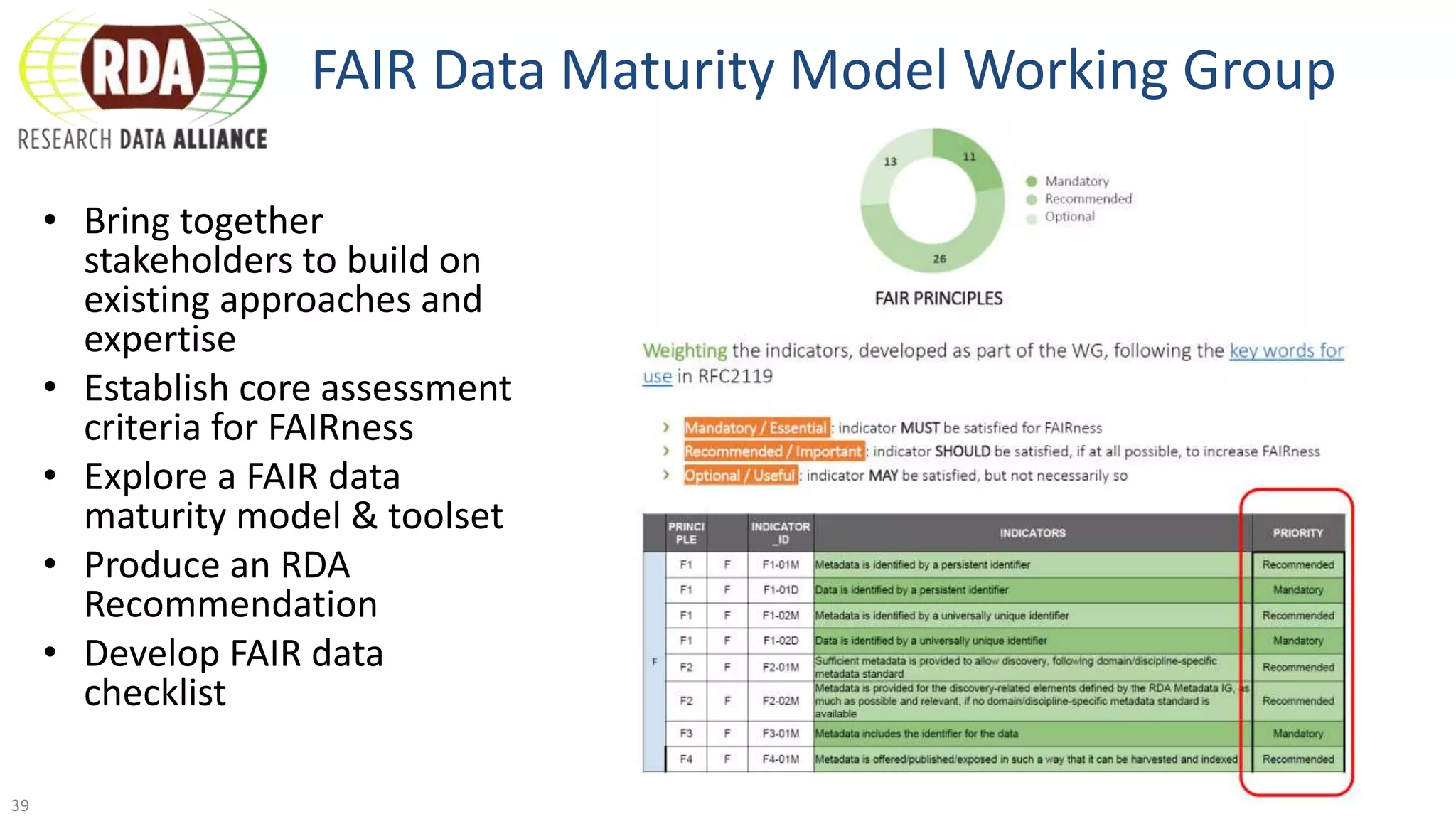
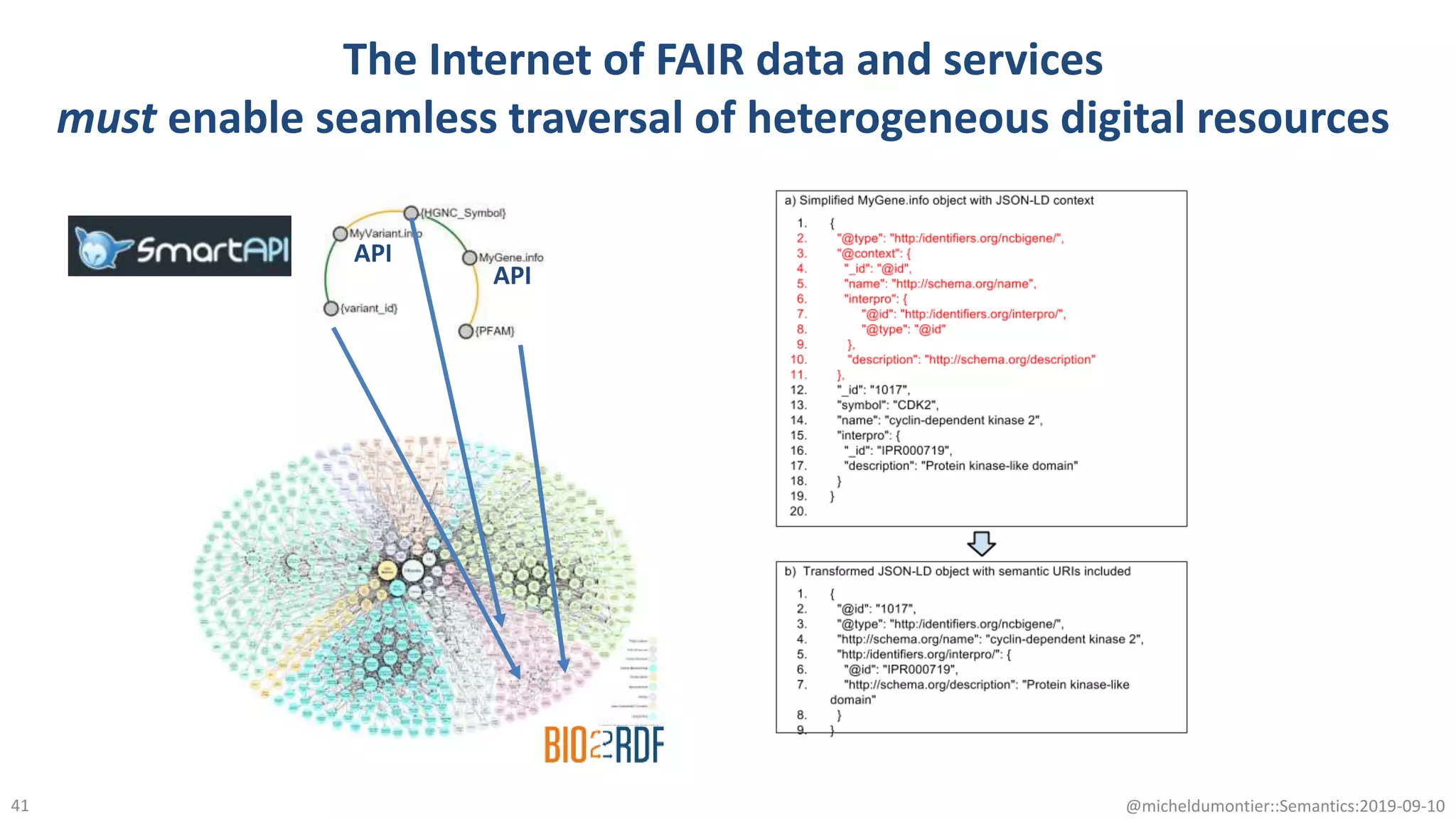
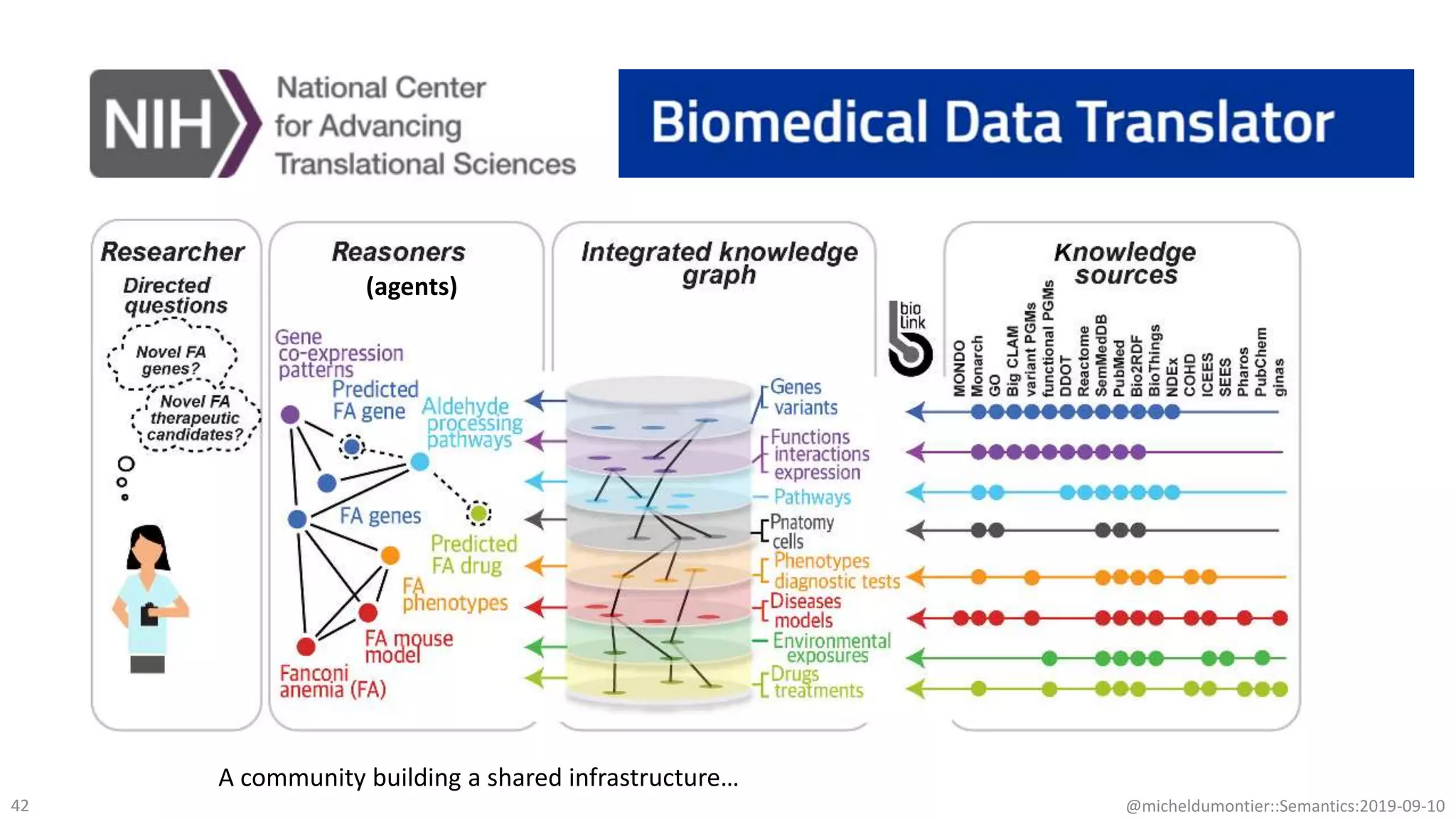
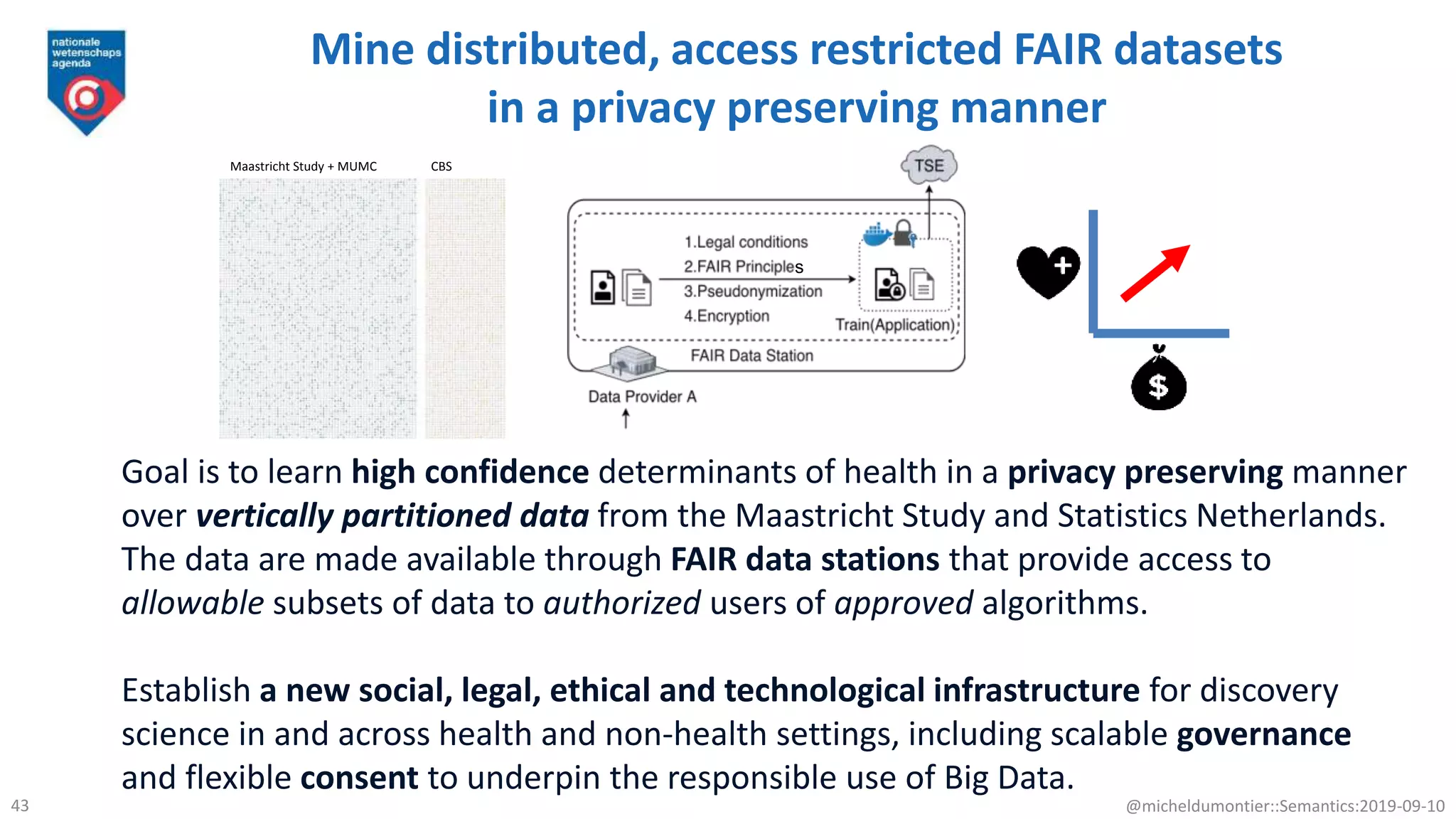
The document discusses the importance of improving data quality and accessibility in biomedical research through the FAIR principles (Findable, Accessible, Interoperable, and Reusable). It highlights challenges such as metadata quality and reproducibility issues, while advocating for a new social and technological infrastructure to facilitate responsible data sharing and collaboration. Ultimately, it underscores the potential of AI and semantic technologies to enhance biomedical discovery by enabling seamless discovery and use of diverse digital resources.