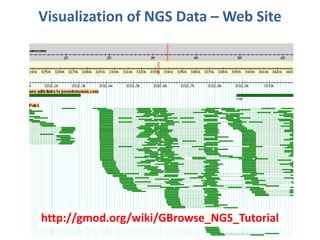
The document summarizes an agenda for a Next Generation Sequencing bioinformatics workshop. It includes:
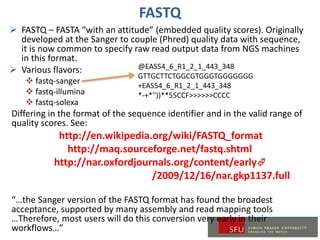
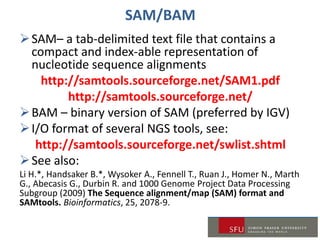
1) A review of common data formats like FASTQ and SAM/BAM used for NGS data as well as tools for working with these formats.
2) An overview of the Picard and FASTX command line tools for processing NGS data.
3) A reintroduction to the Galaxy platform for analyzing NGS data through an online tutorial.
4) Suggested steps for a typical NGS data analysis workflow including quality control, assembly, and mapping.