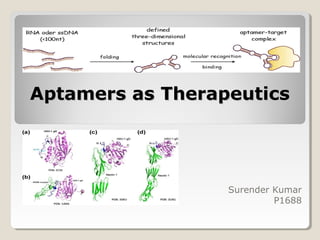
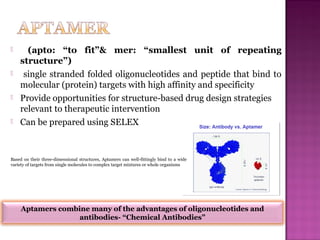
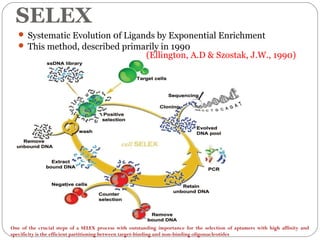
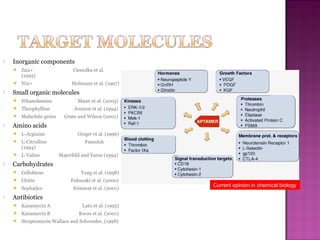
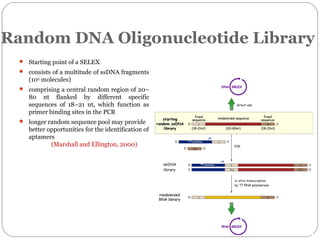
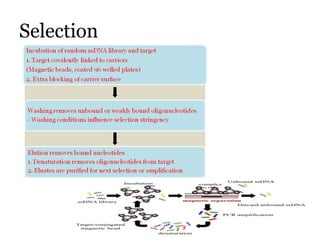
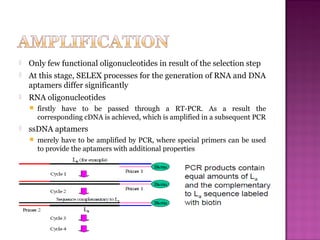
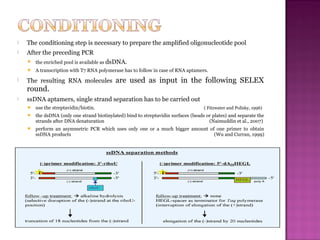
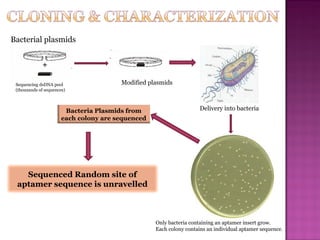
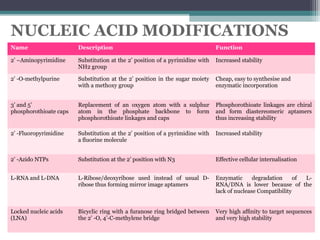
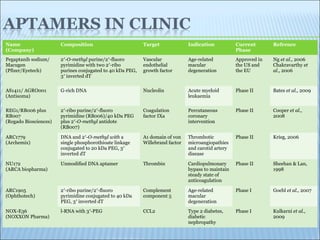
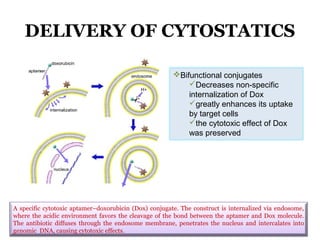
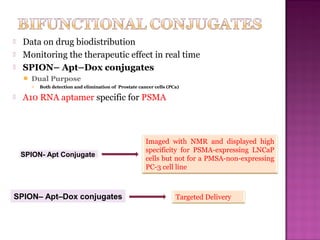
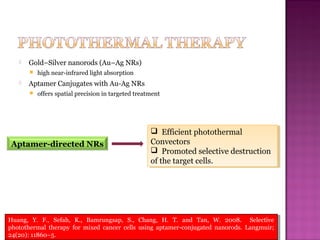
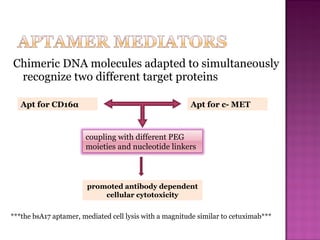
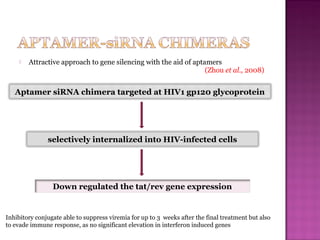
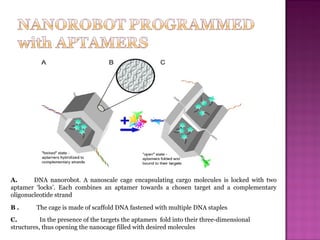
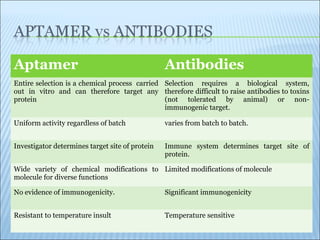
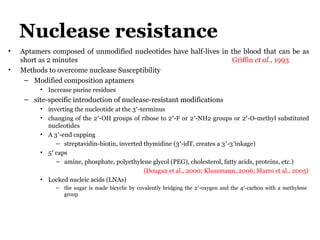
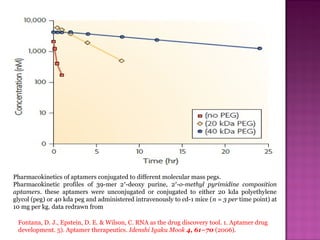
The document discusses the use of aptamers as therapeutic agents, highlighting their high affinity and specificity in binding to molecular targets, akin to 'chemical antibodies.' It elaborates on the SELEX process for aptamer selection and various modifications to enhance their stability and efficacy, as well as their applications in drug development and targeted therapy. Overall, it suggests that aptamers are emerging as viable alternatives to monoclonal antibodies in the therapeutic landscape.