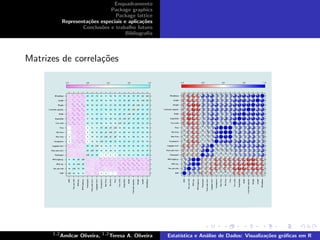
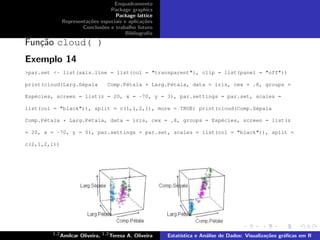
This document provides examples of various plotting functions in R including plot(), boxplot(), hist(), pairs(), barplot(), densityplot(), dotplot(), histogram(), xyplot(), cloud(), and biplot/triplot. Functions are demonstrated using built-in datasets like iris and by plotting variables against each other to create scatter plots, histograms, and other visualizations.


























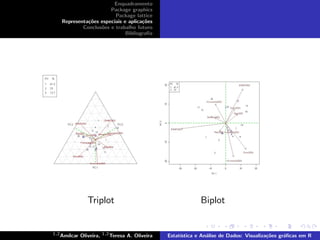
![biplot e triplot)
> library(agricolae)
> library(klaR)
> data(Oat2)
> startgraph
> biplot
> model<- AMMI(Oat2[,1], Oat2[,2], Oat2[,3], Oat2[,4],xlim=c(-35,20),ylim=c(-20,20),
graph="biplot")
> model<- AMMI(Oat2[,1], Oat2[,2], Oat2[,3], Oat2[,4],xlim=c(-35,20),ylim=c(-20,20),
graph="biplot",number=FALSE)
> triplot
> model<- AMMI(Oat2[,1], Oat2[,2], Oat2[,3], Oat2[,4],graph="triplot")](https://image.slidesharecdn.com/joclad2010d-120223193436-phpapp02/85/Joclad-2010-d-28-320.jpg)