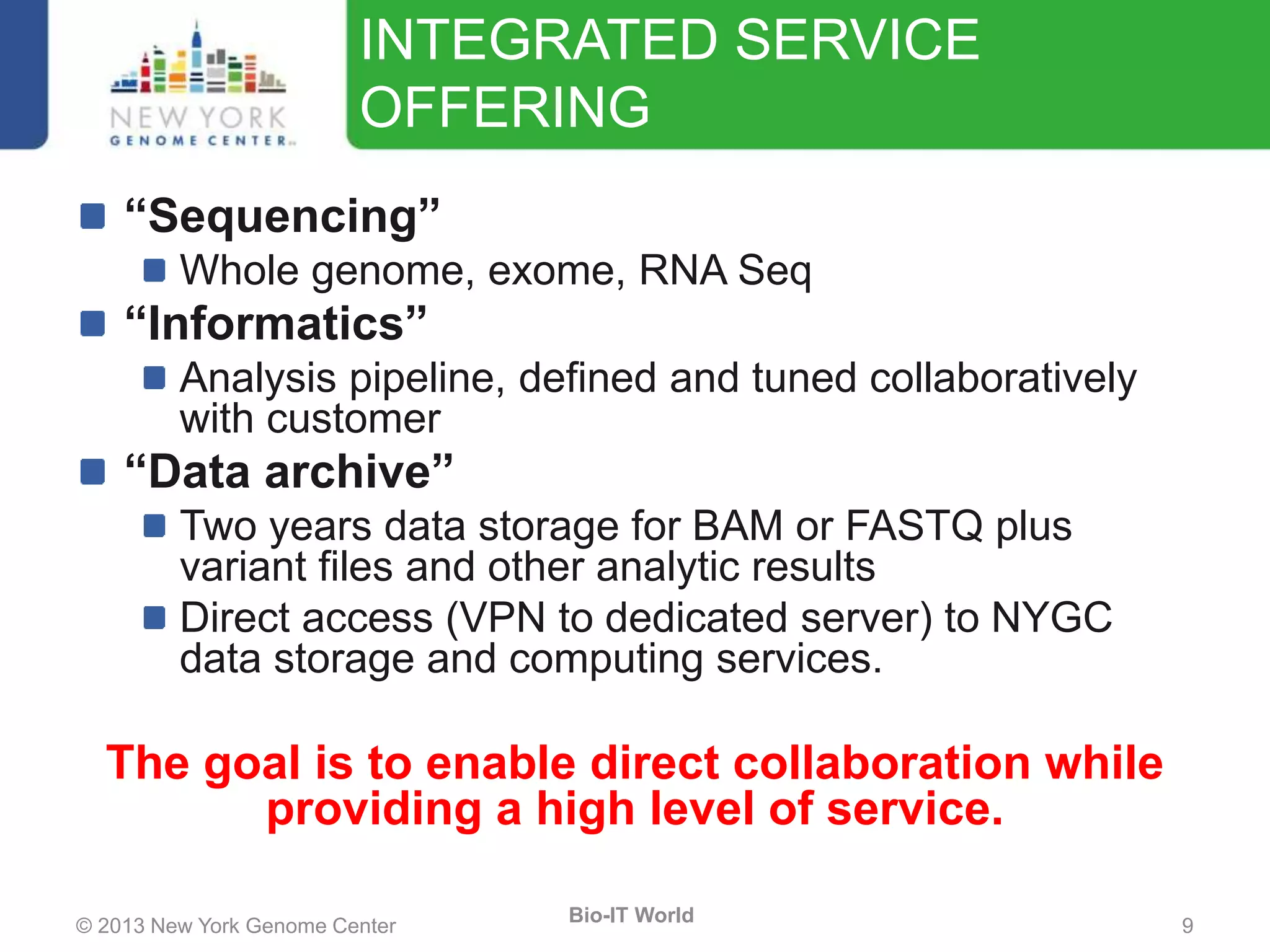
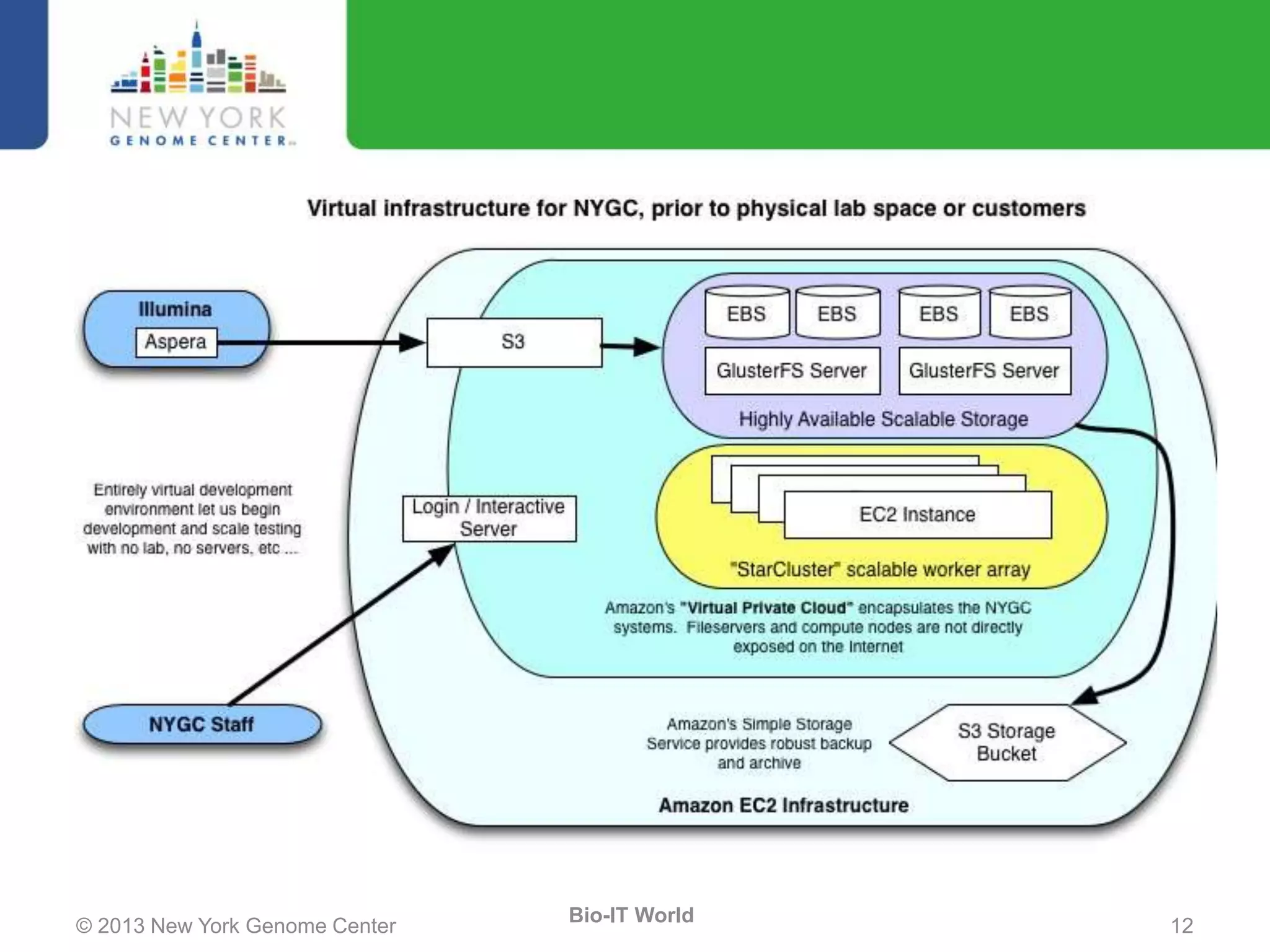
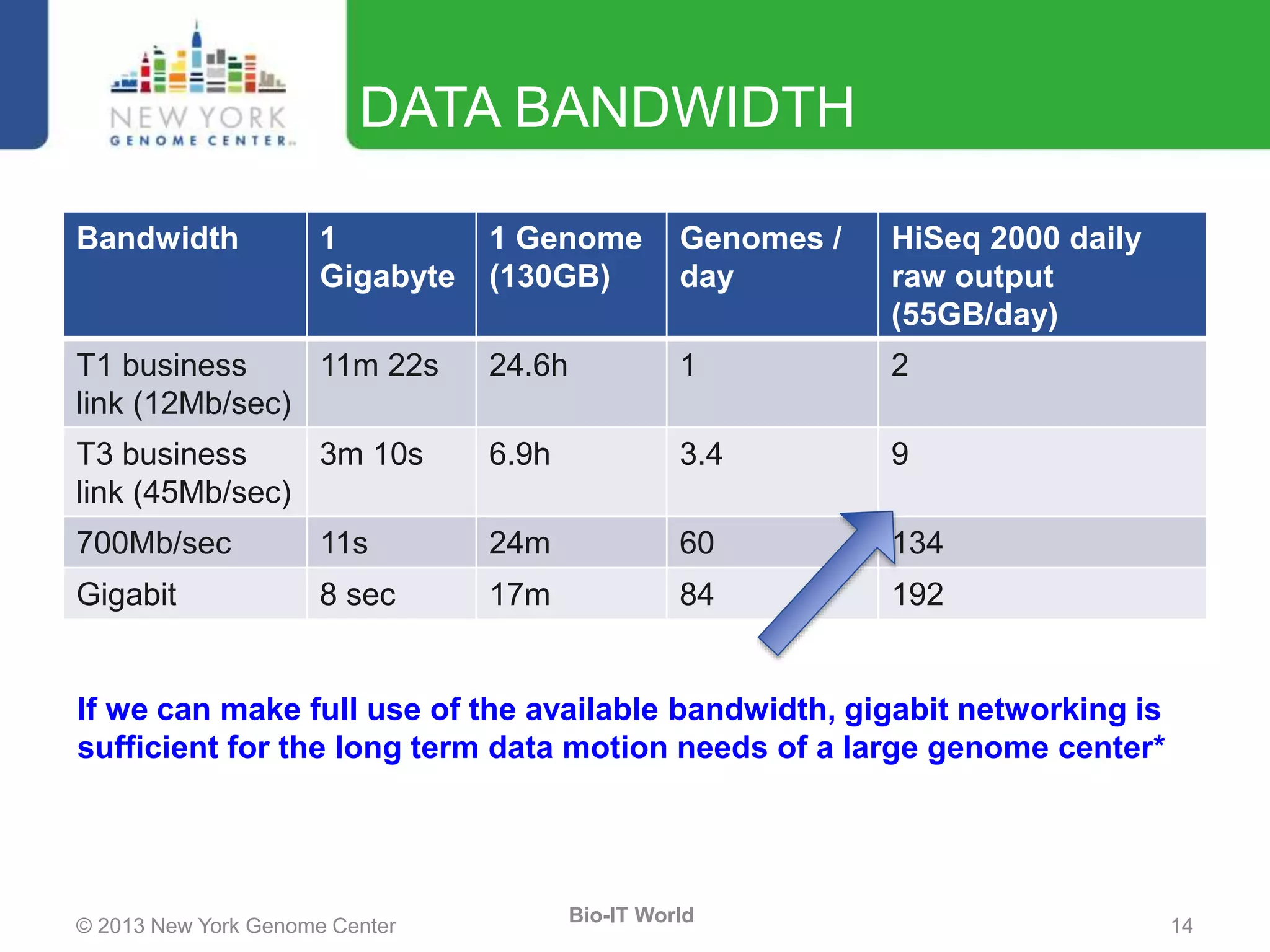
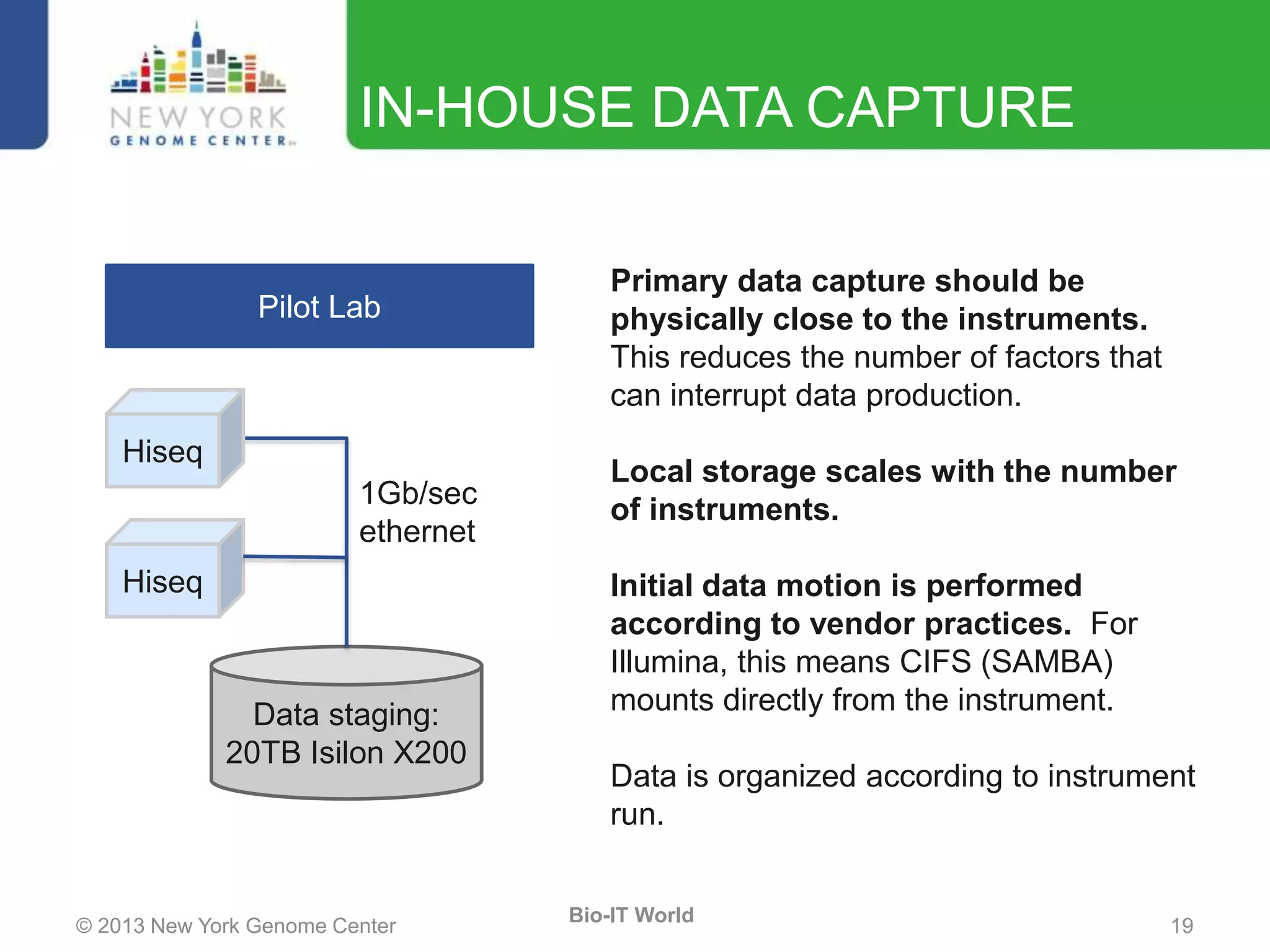
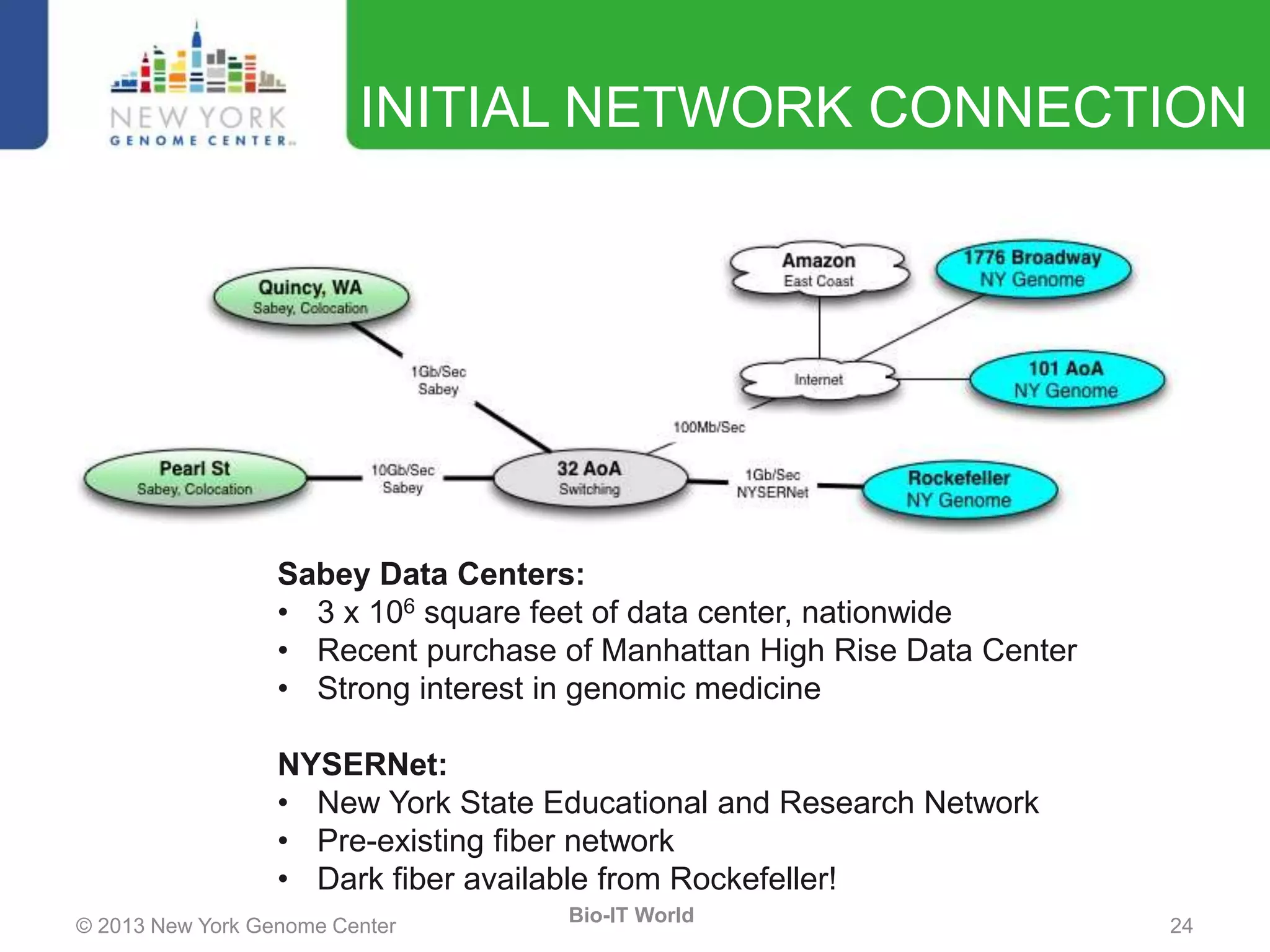
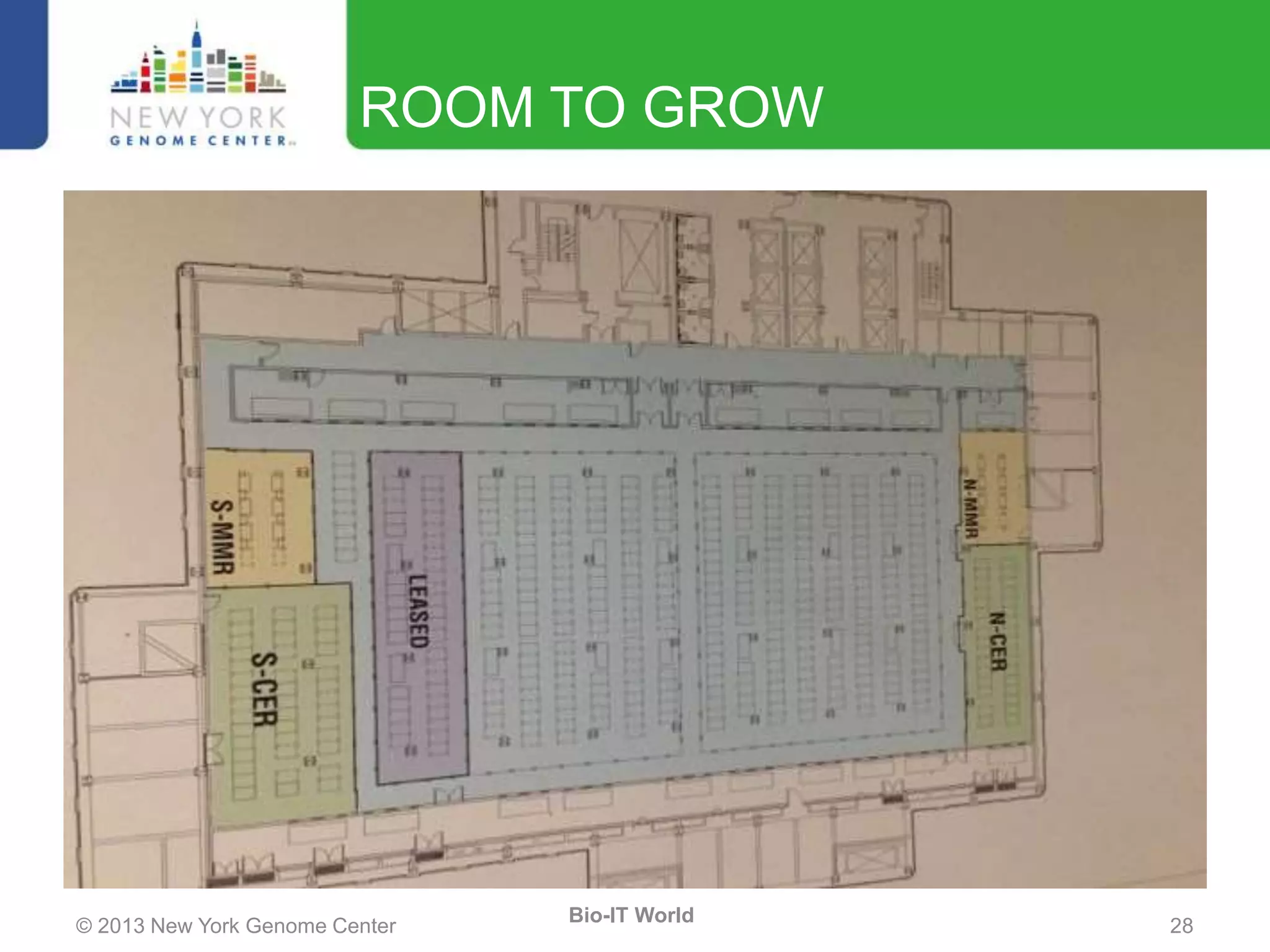
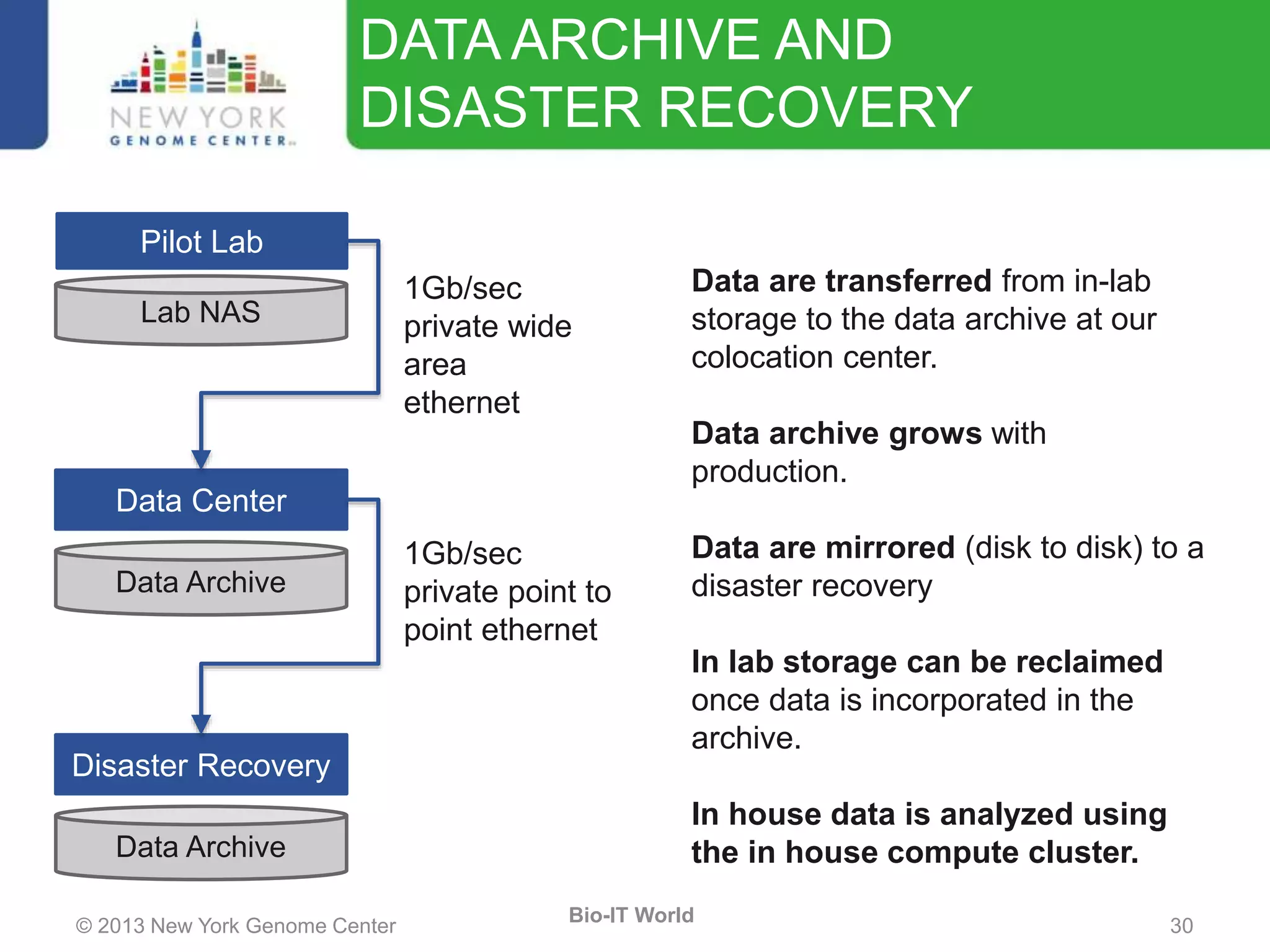
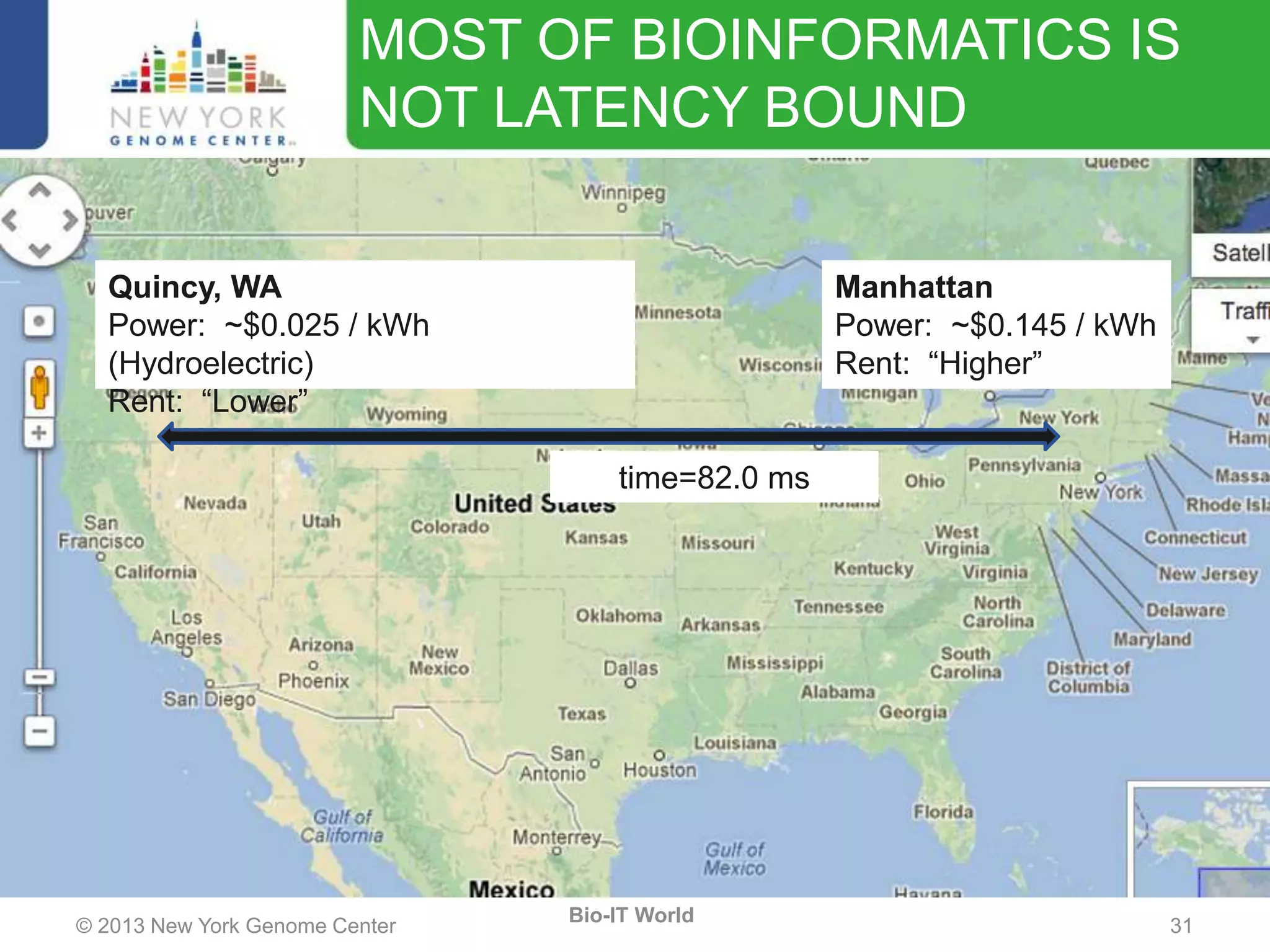
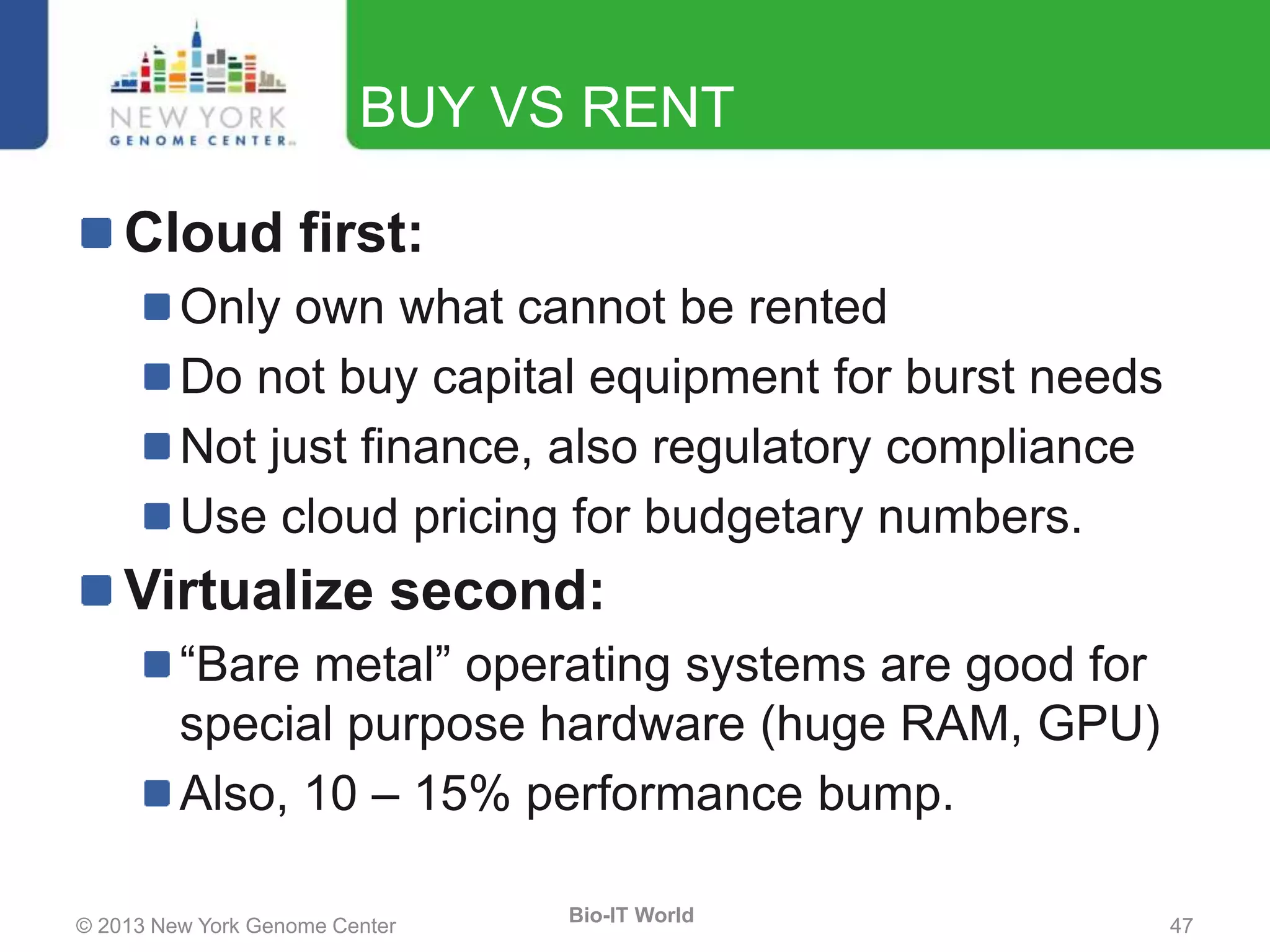
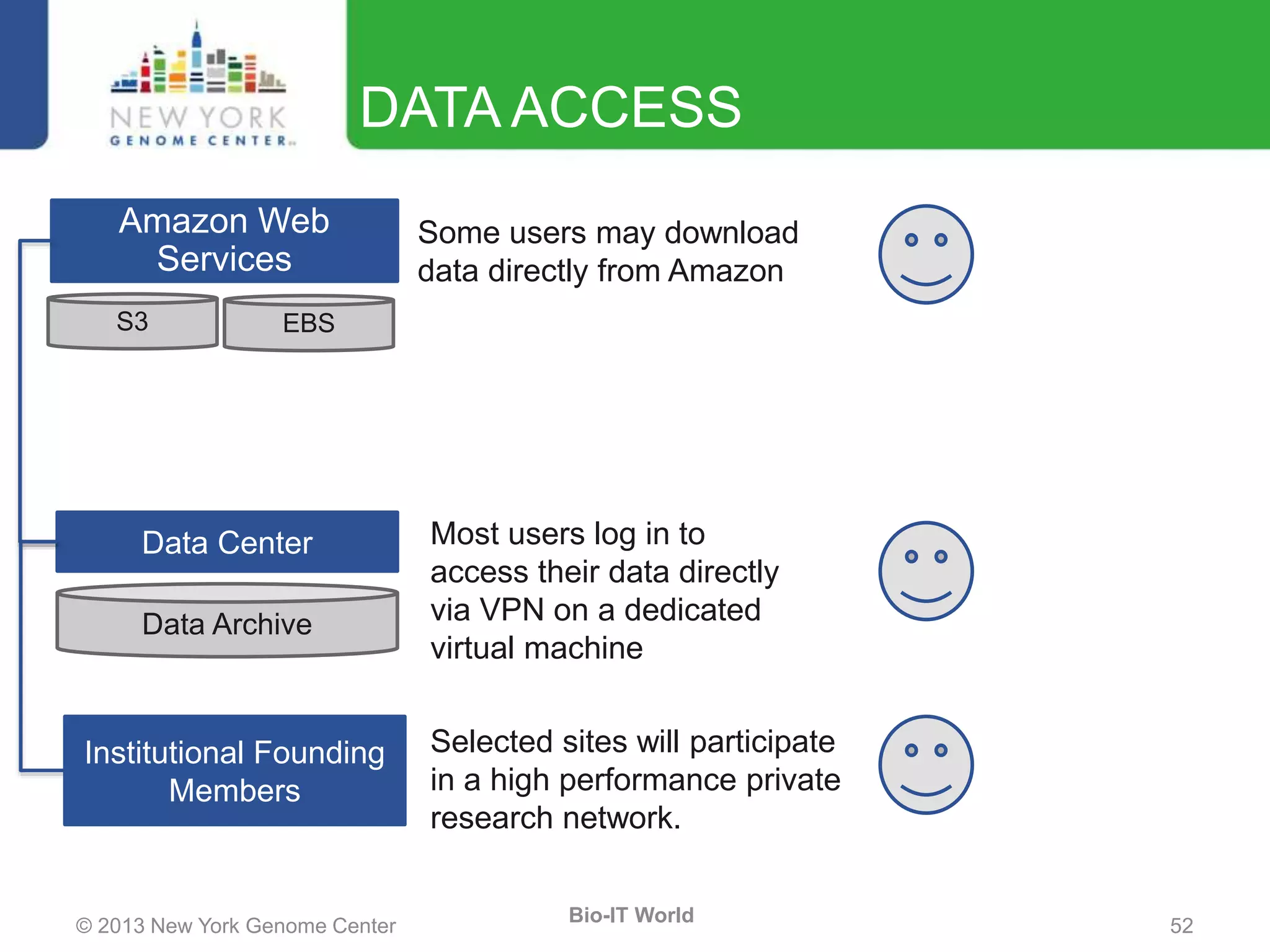
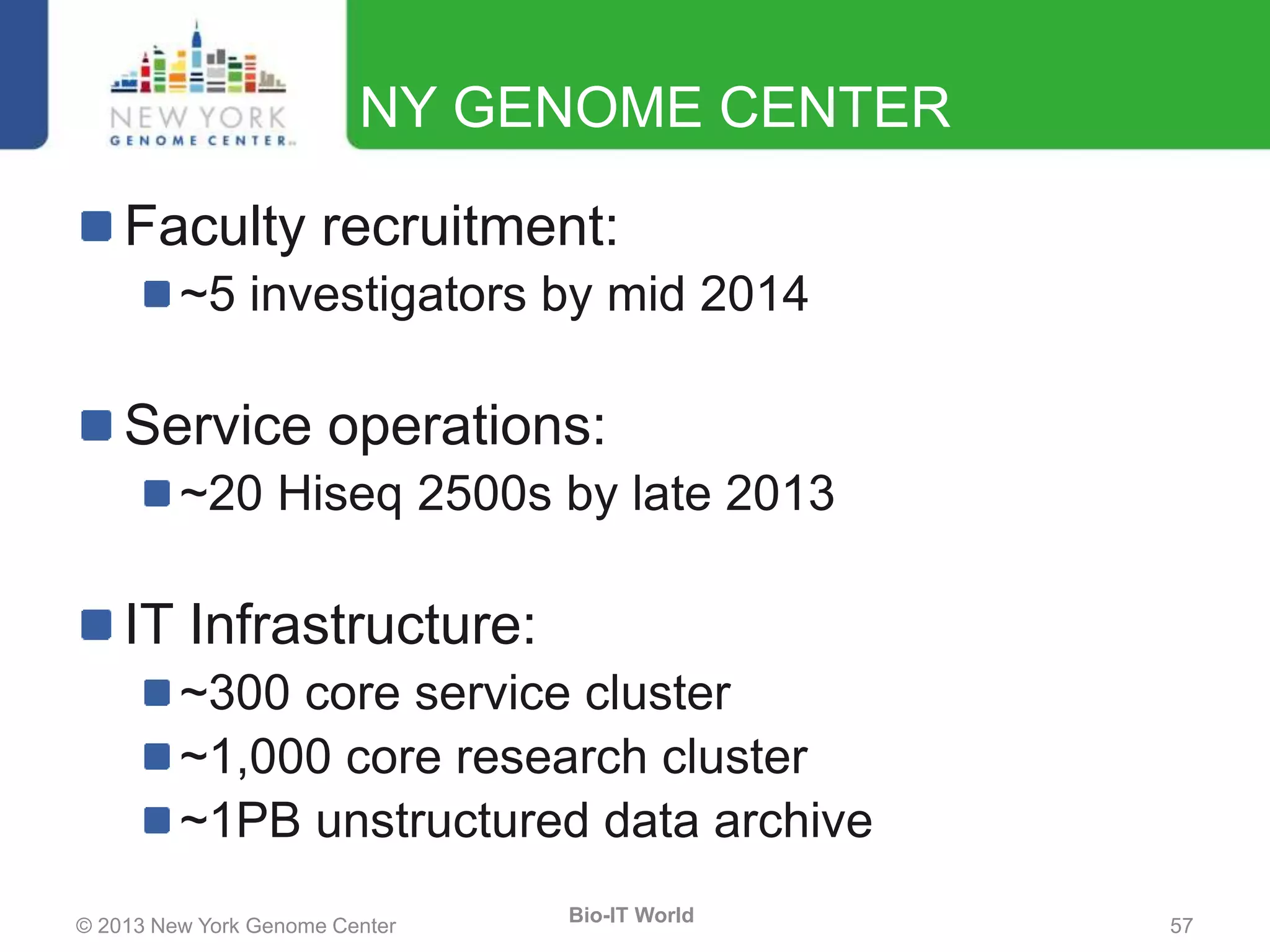
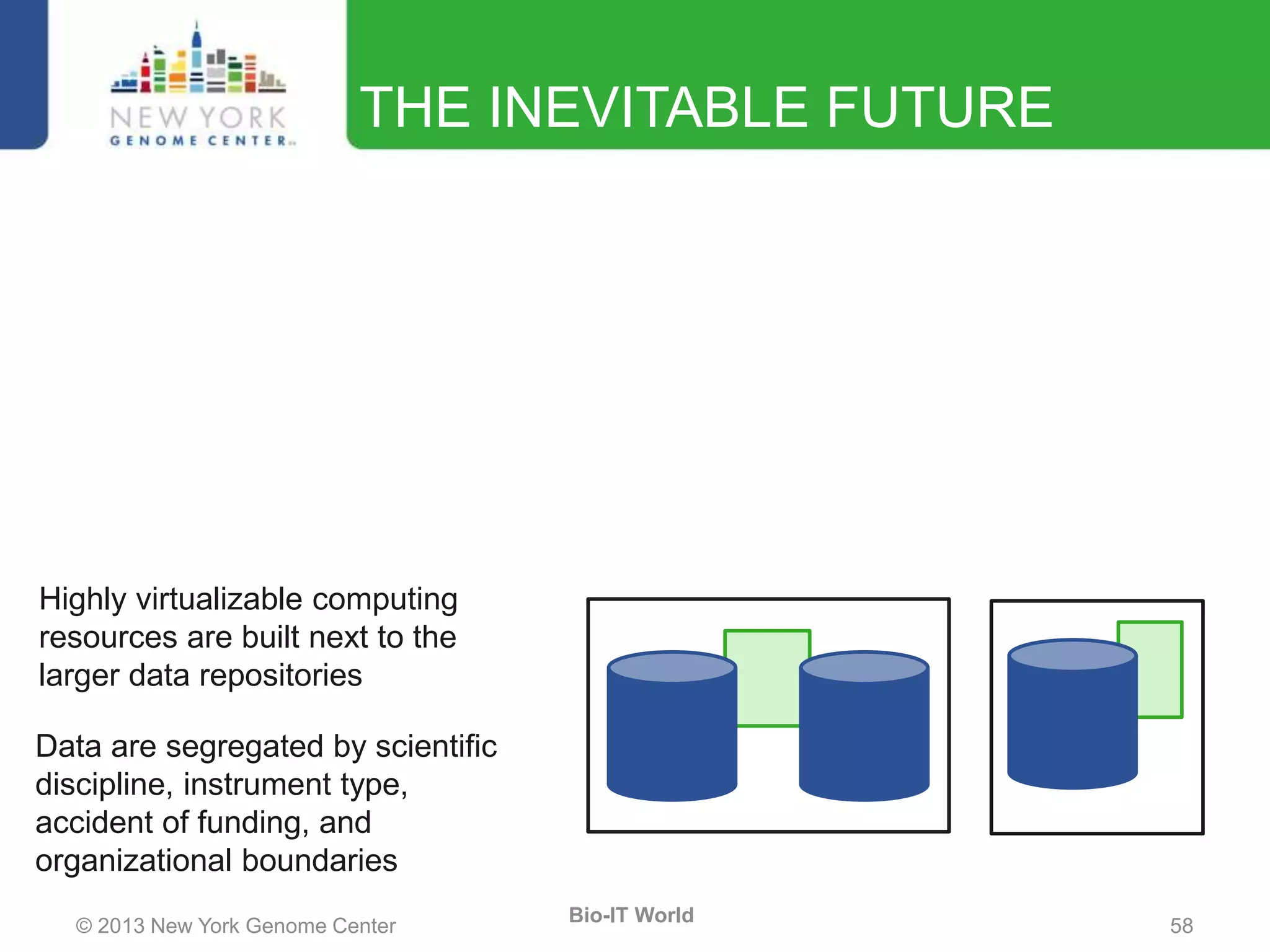
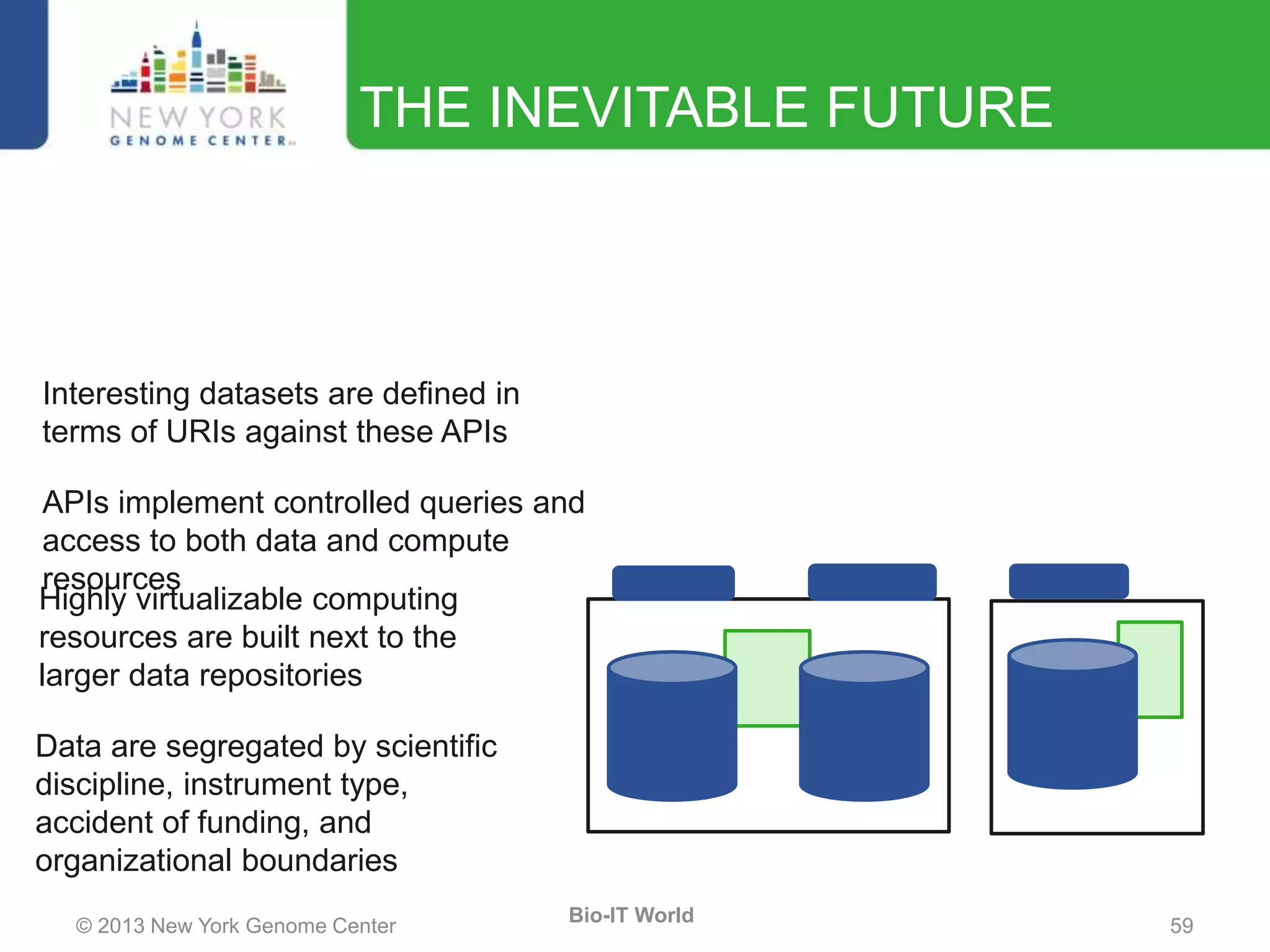
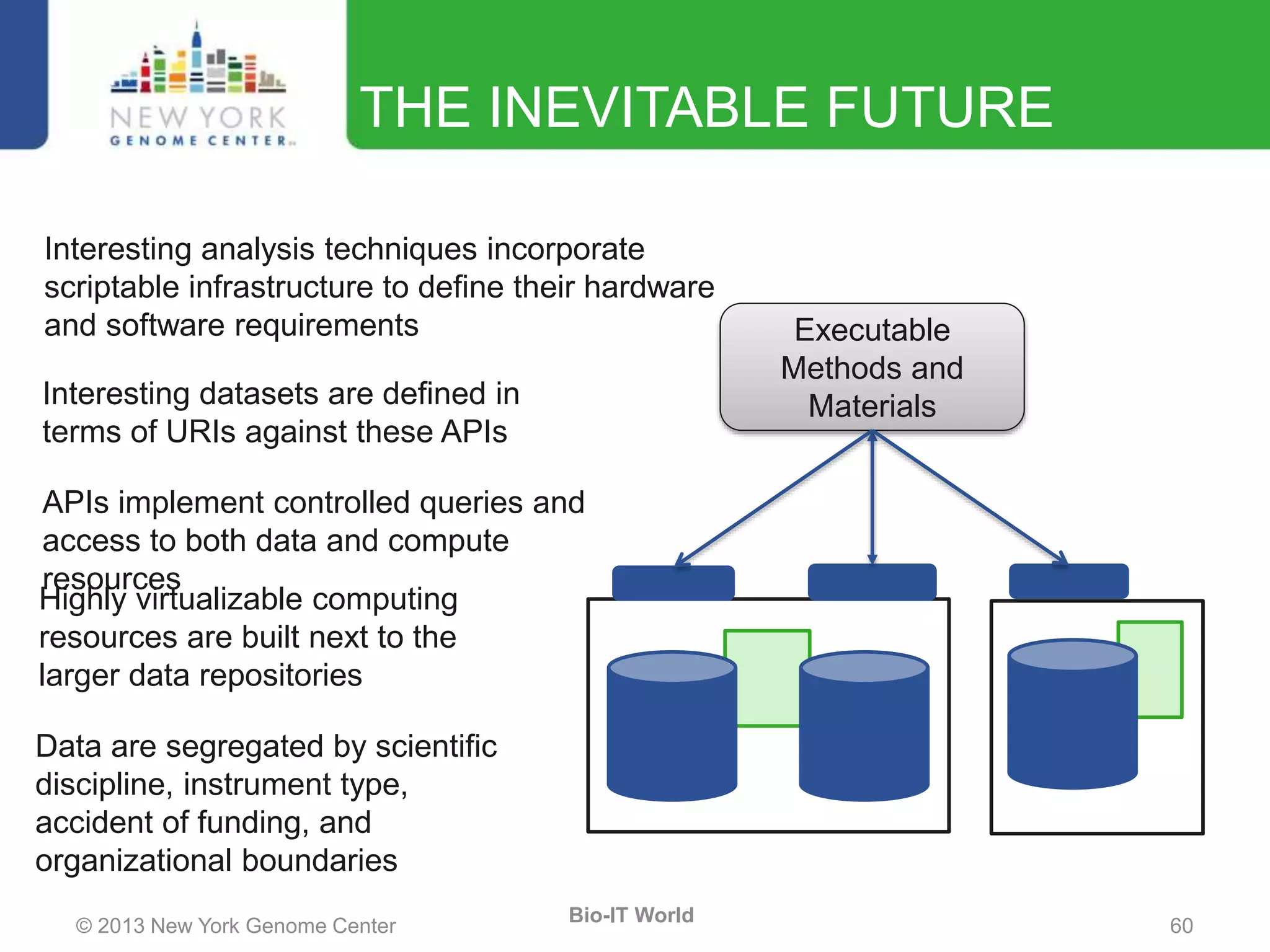
The document outlines the IT infrastructure and operations of the New York Genome Center, emphasizing the importance of centralized resources for sequencing, bioinformatics, and data storage. It discusses the future of data management, highlighting the necessity of flexible APIs, cloud computing, and effective data archives to enable collaboration and scientific success. The report also details the physical setup of its facilities, network connectivity, and disaster recovery measures to support large-scale genomic research.