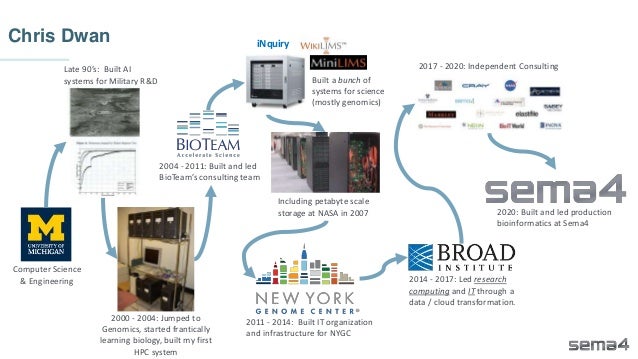
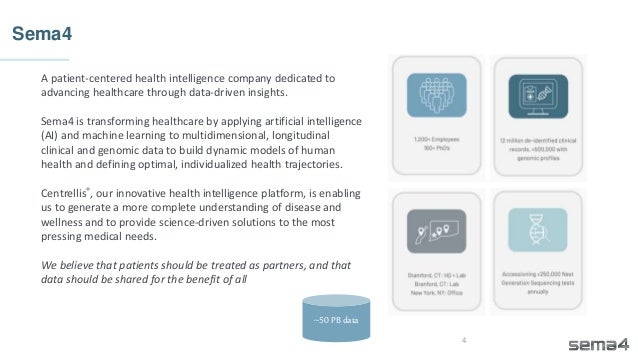
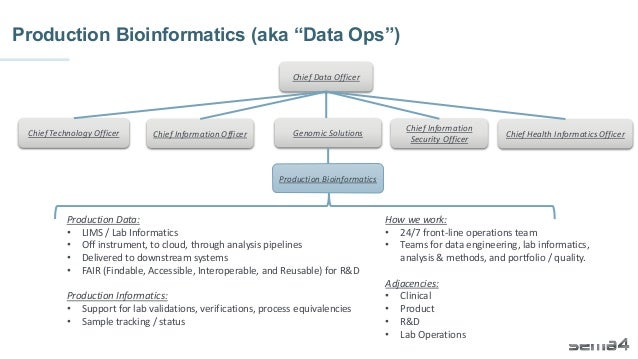
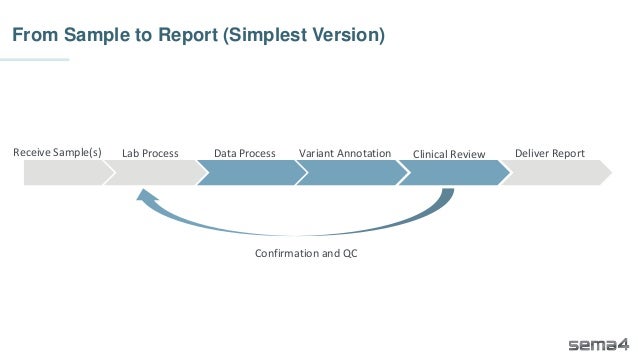
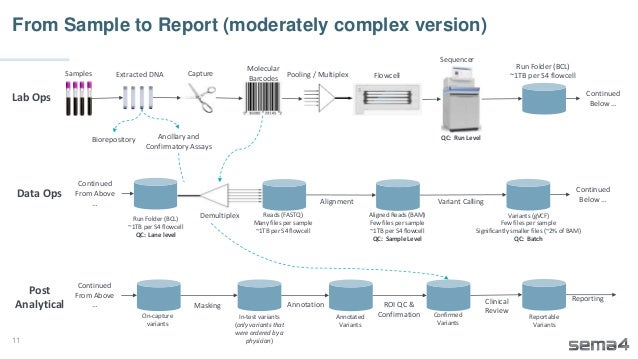
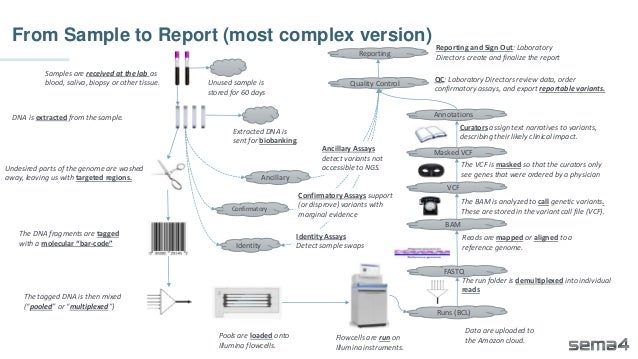
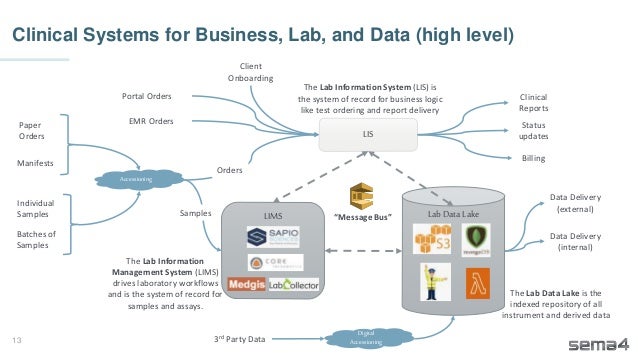
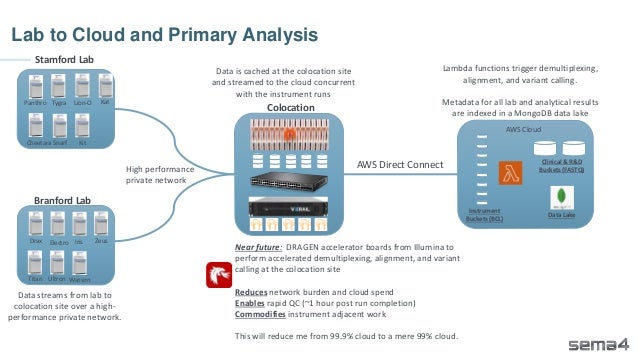
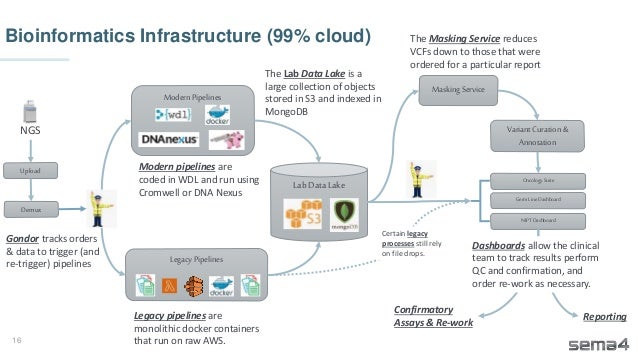
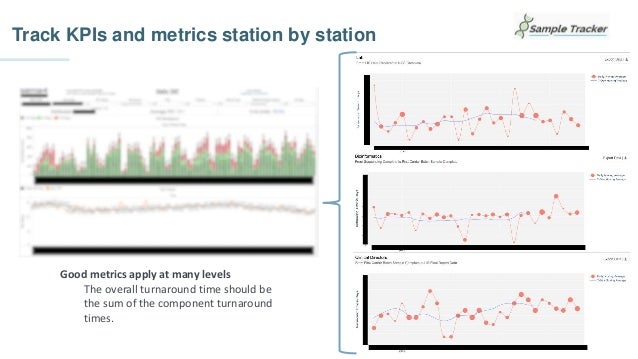
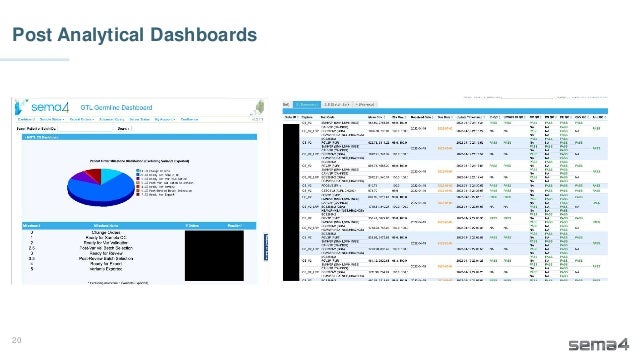
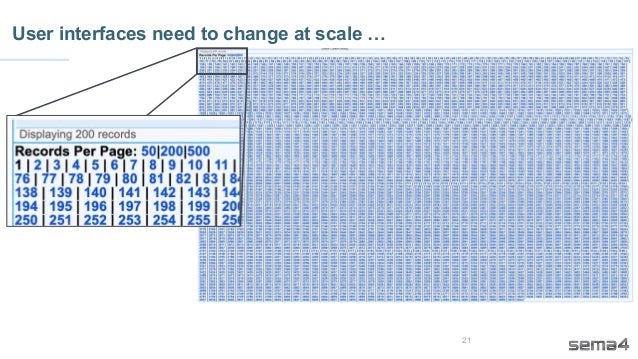
Chris Dwan emphasizes the importance of production bioinformatics and the integration of data-driven insights in healthcare at Sema4. Sema4 leverages AI and machine learning to transform clinical and genomic data into personalized health trajectories, offering various genomic tests and data platforms. The document details operational methodologies, data management techniques, and a focus on maintaining high quality and accountability in bioinformatics workflows.