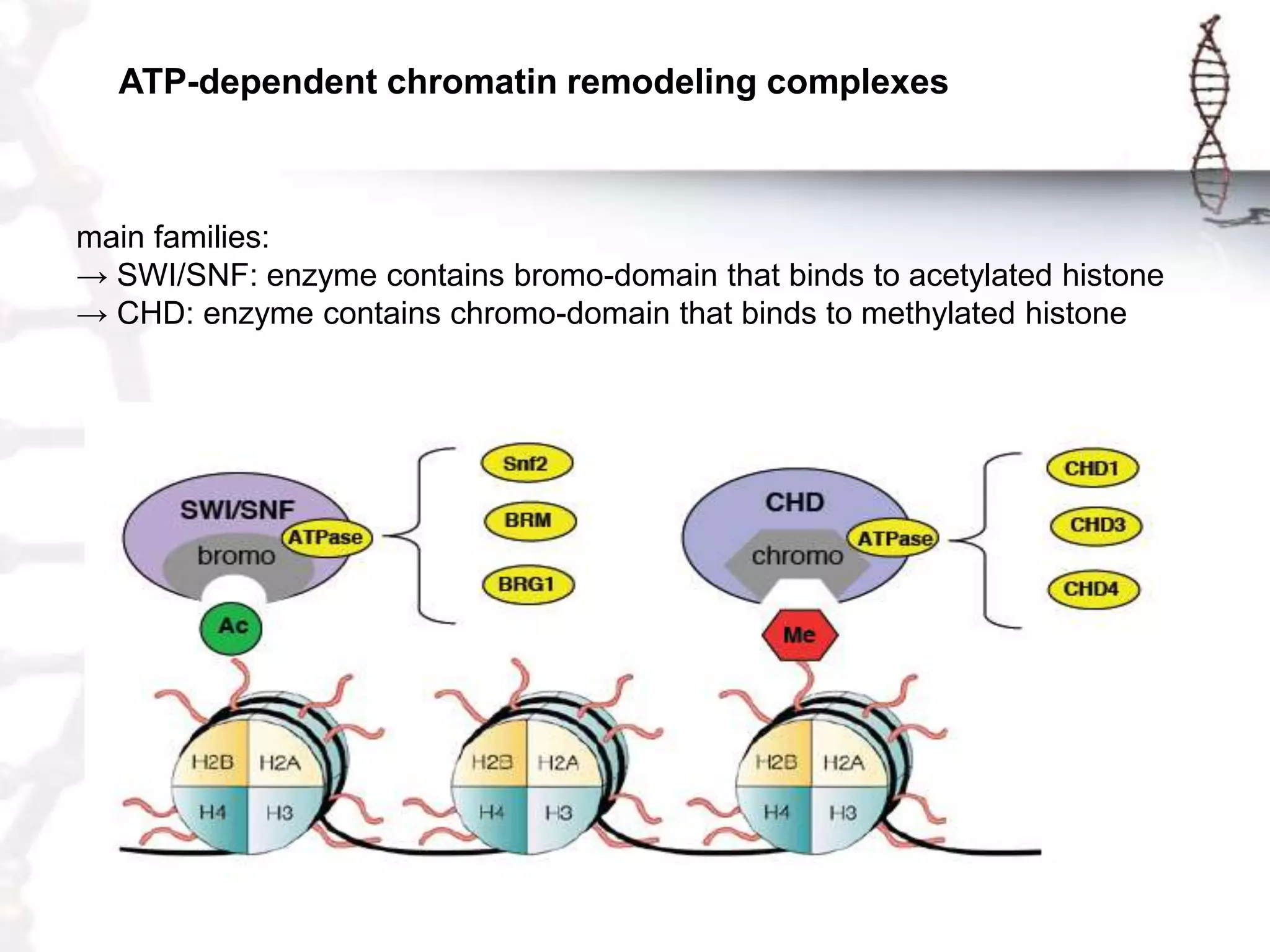
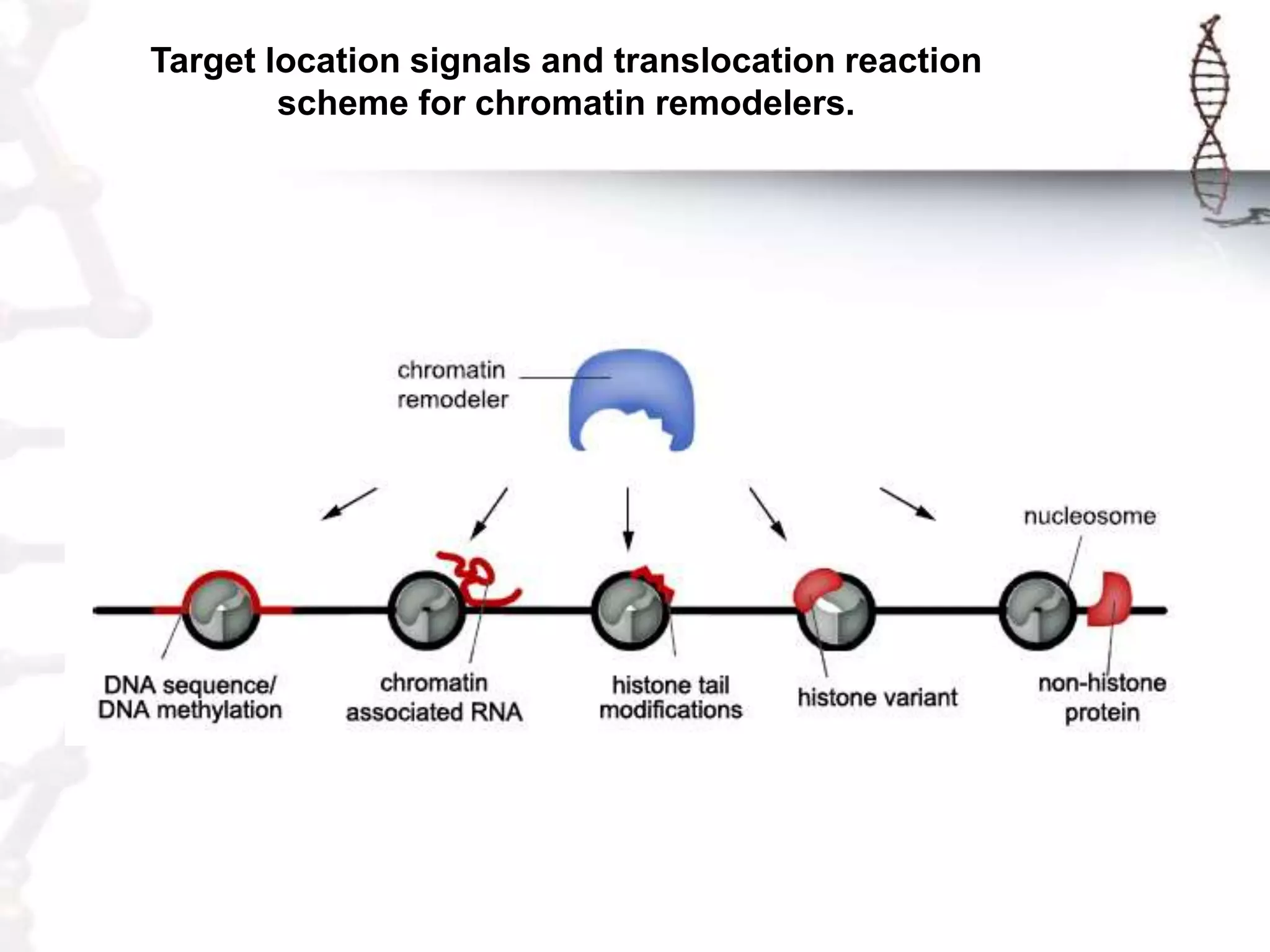
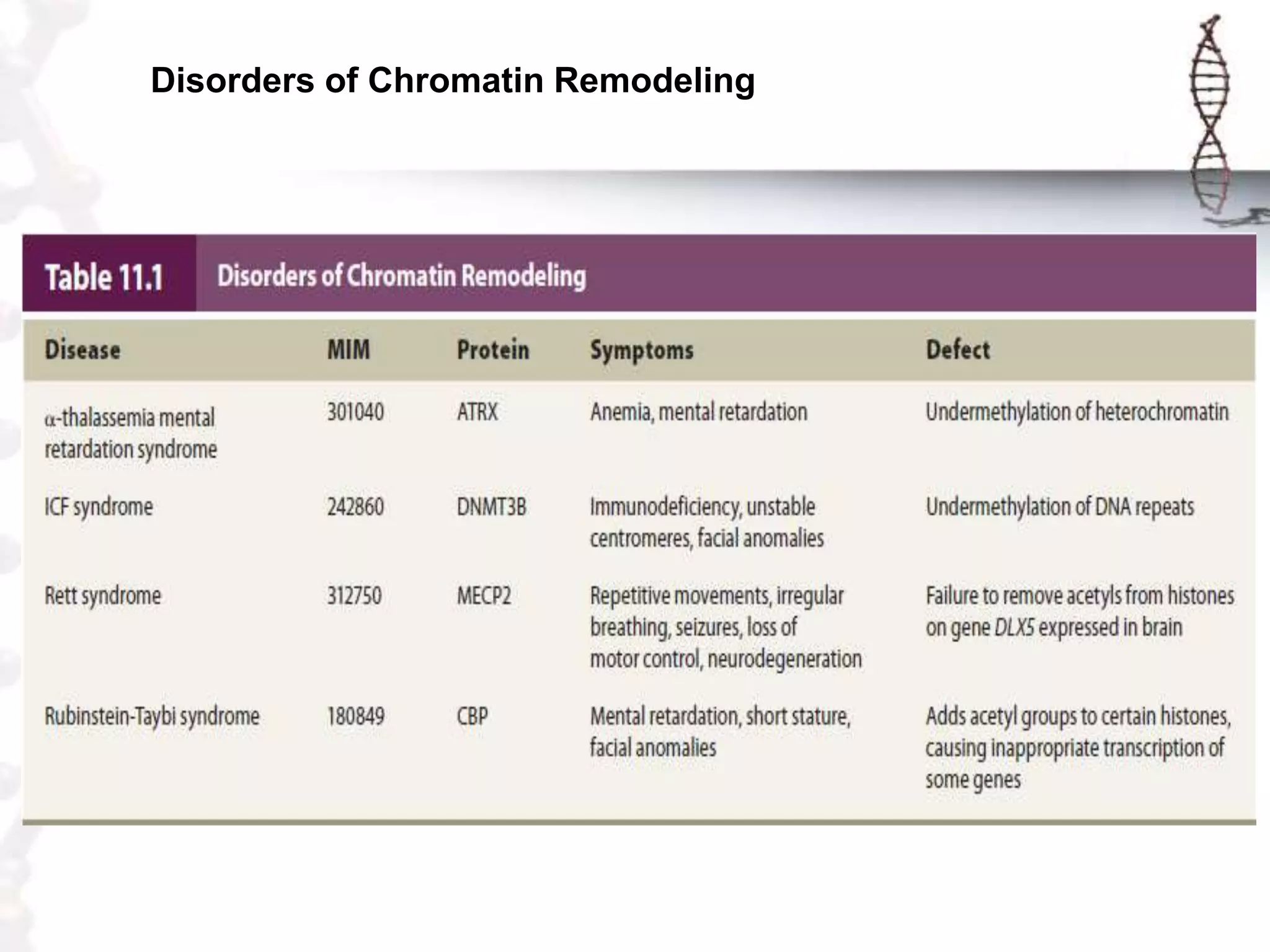
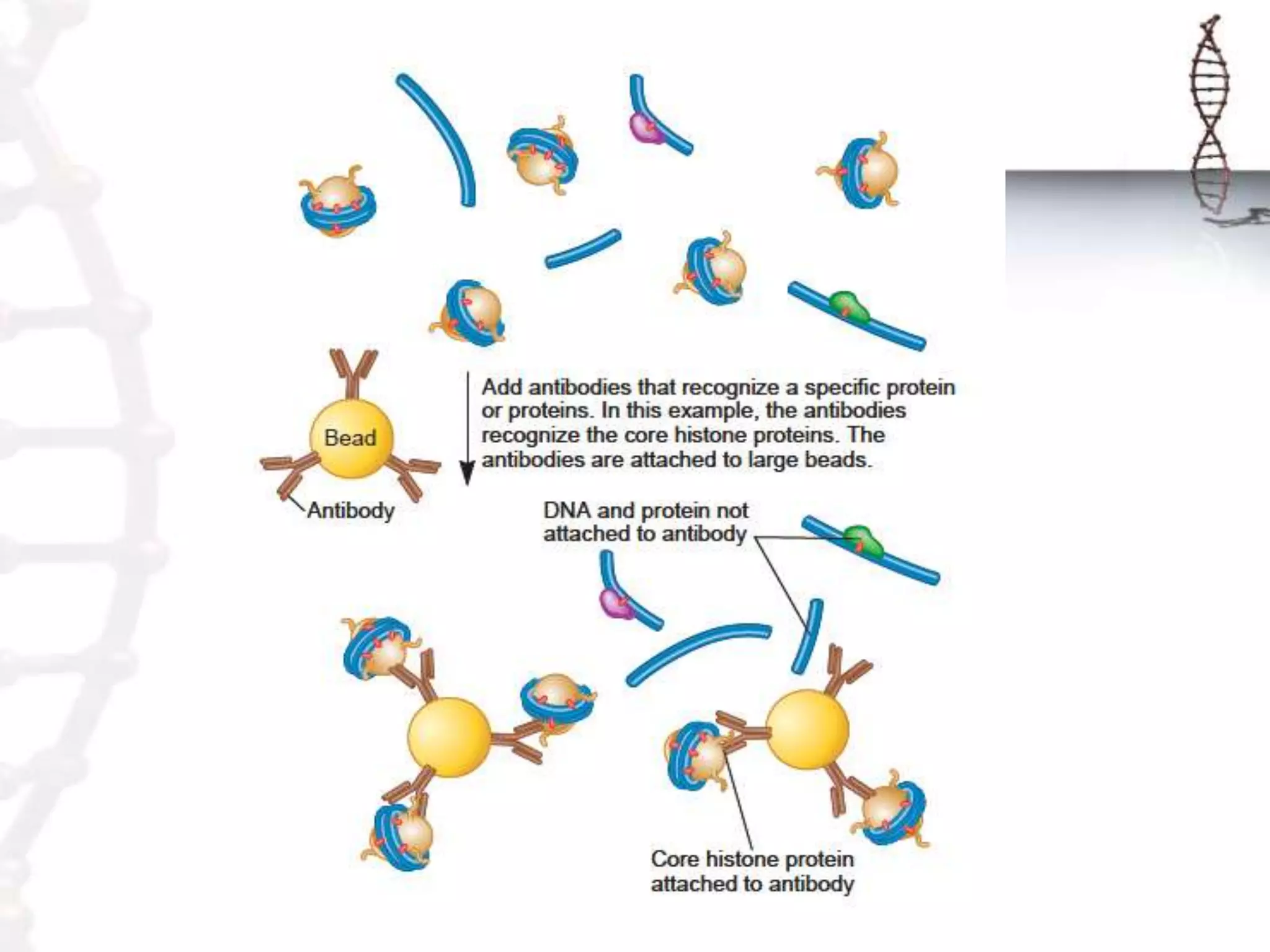
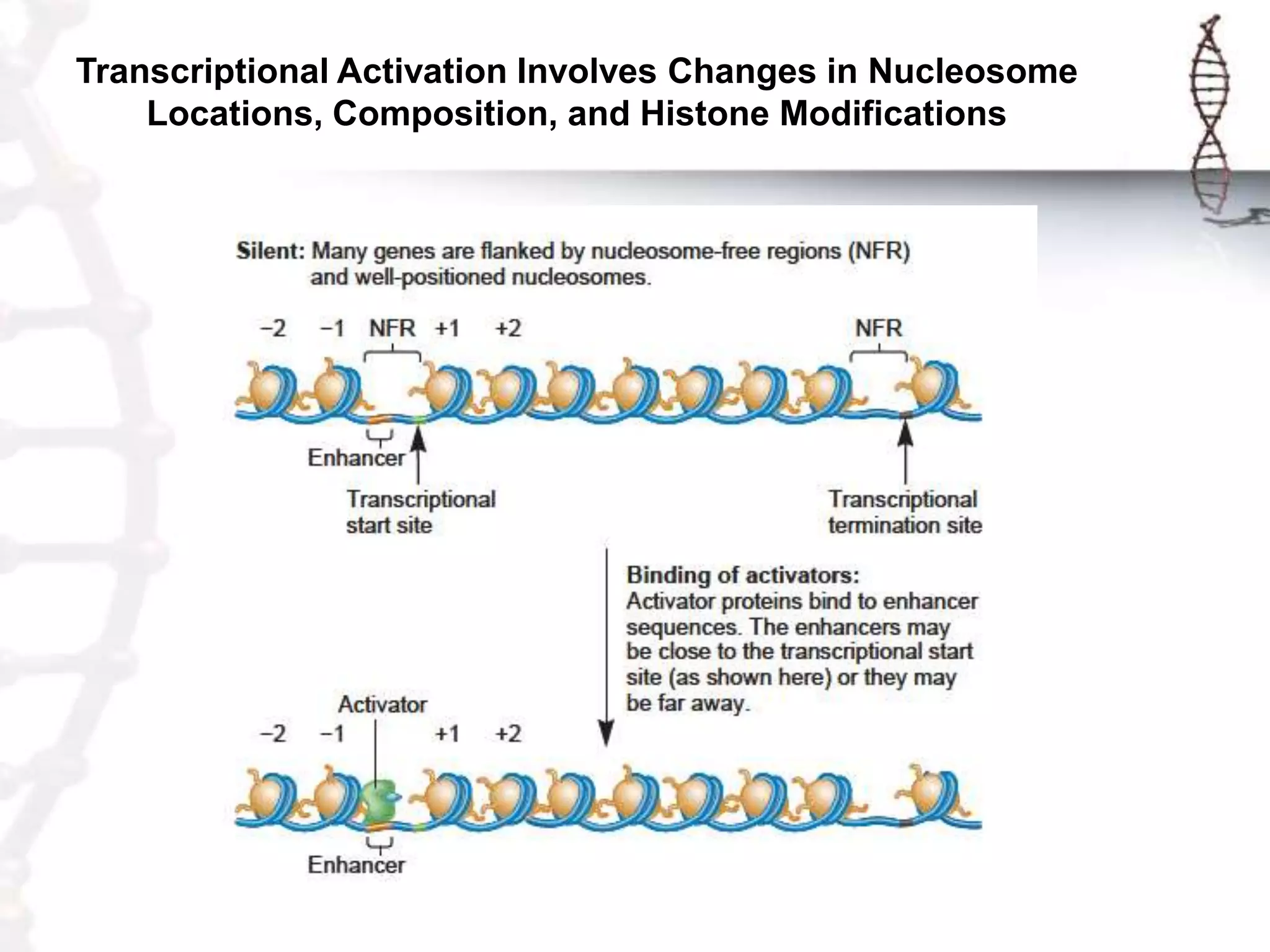
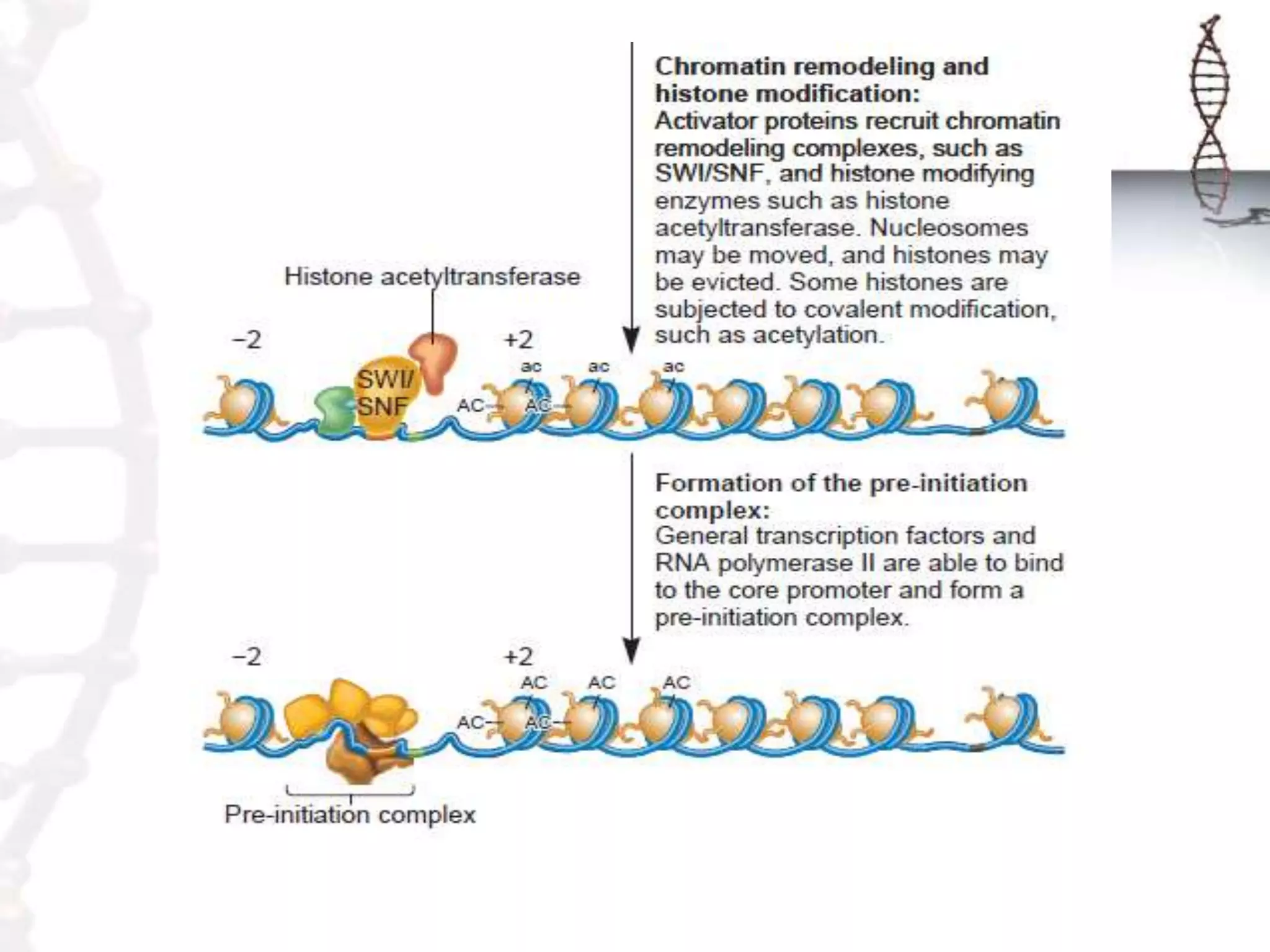
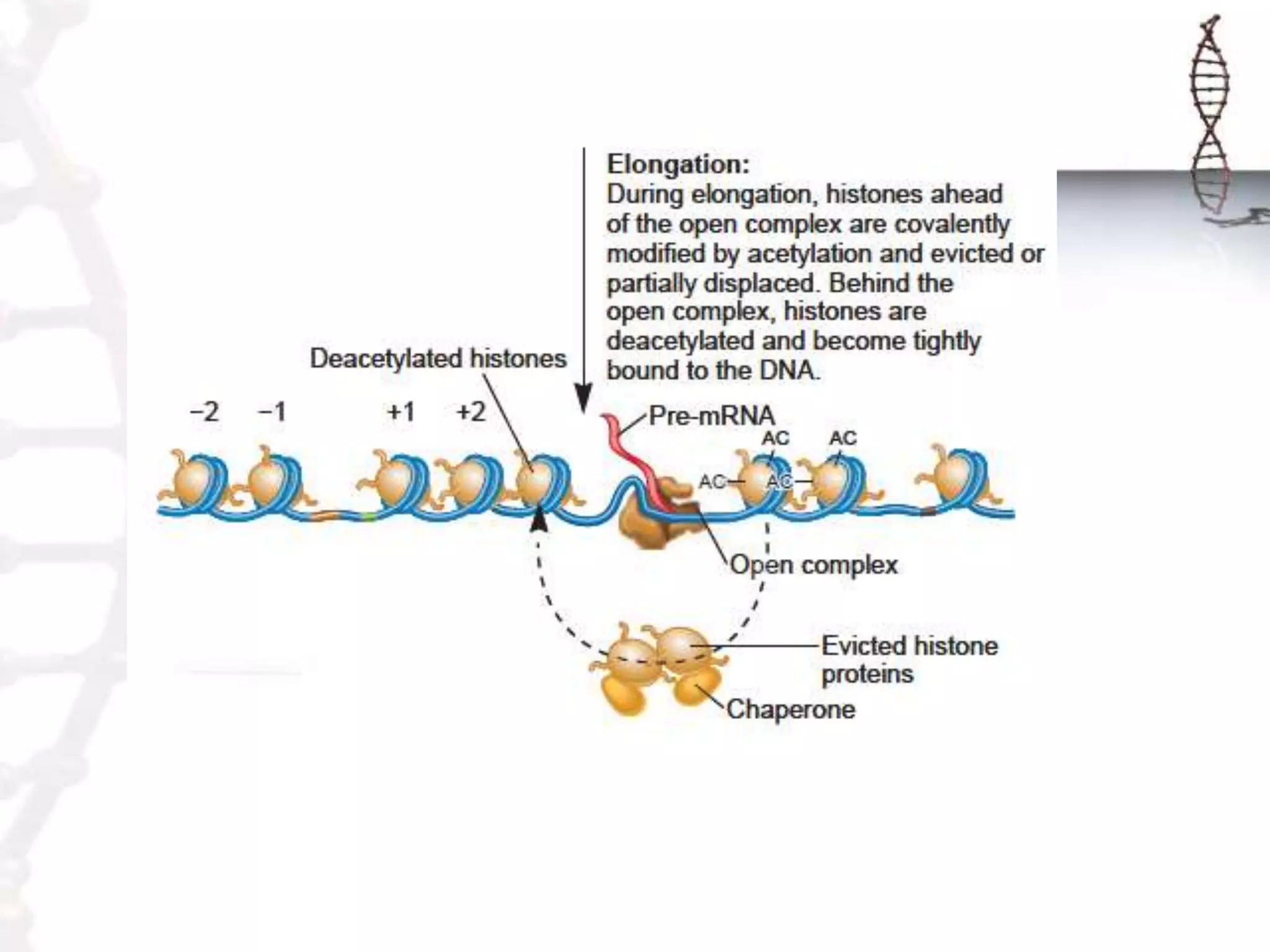
Chromatin remodeling refers to dynamic changes in chromatin structure that occur in cells. There are several classes of chromatin remodeling complexes that use ATP hydrolysis to alter nucleosome positioning in different ways: 1) Nucleosome sliding, 2) Ejection of histone dimers or octamers, and 3) Replacement of core histones with histone variants. Disorders can result if chromatin remodeling is disrupted. Techniques like chromatin immunoprecipitation sequencing are used to study chromatin structure.