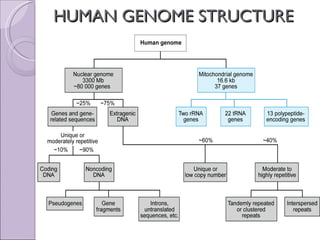
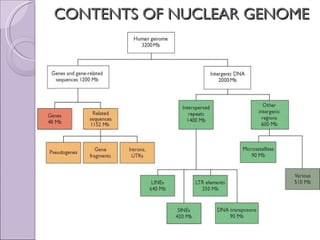
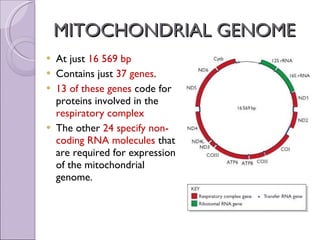
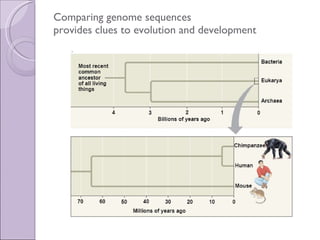
The document discusses several topics related to genome evolution, including:
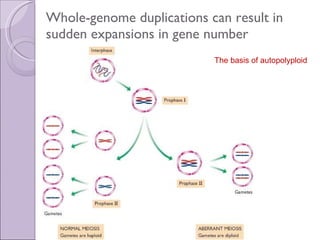
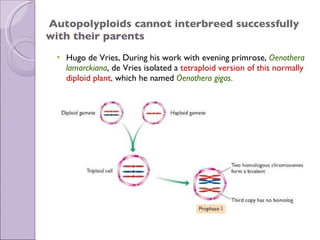
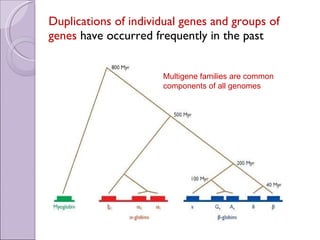
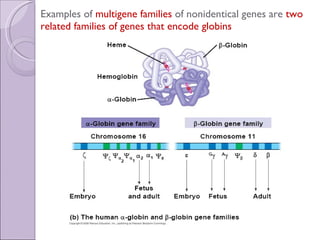
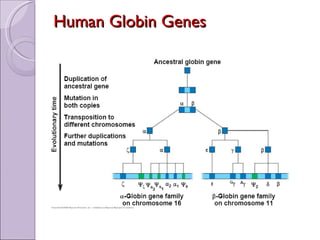
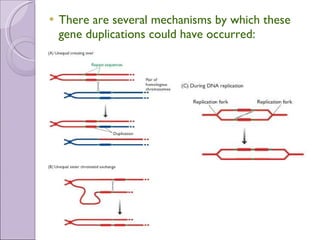
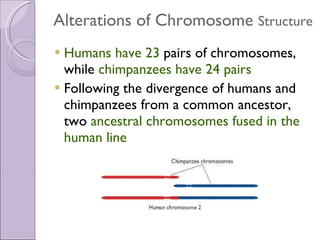
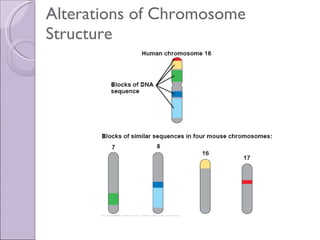
1. Genome evolution involves the acquisition of new genes through gene duplication within genomes or lateral gene transfer between species. Gene duplication can occur at the whole genome level or for individual or groups of genes.
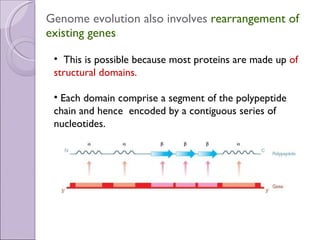
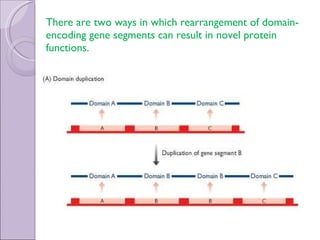
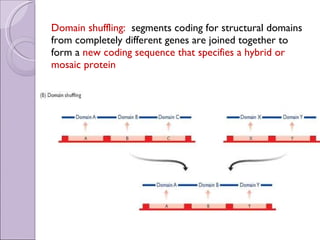
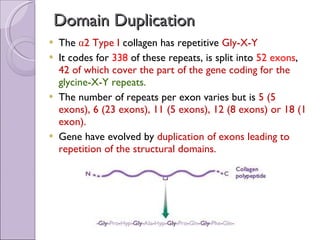
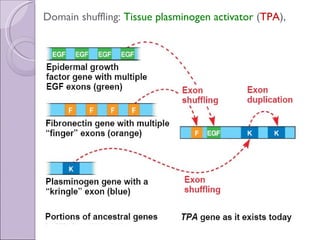
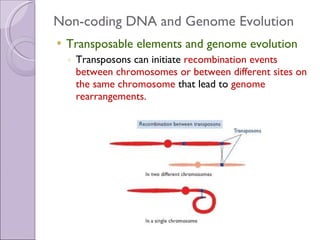
2. Genome evolution also involves rearrangement of existing genes through domain shuffling, where gene segments are joined to form new proteins, or domain duplication where repetitive domains evolve new functions.
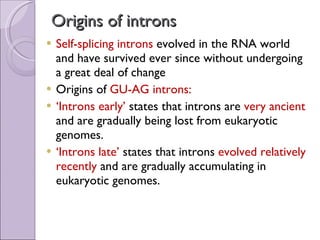
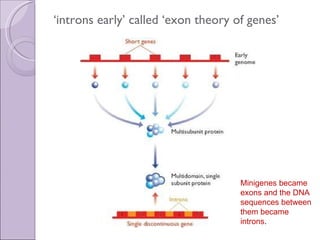
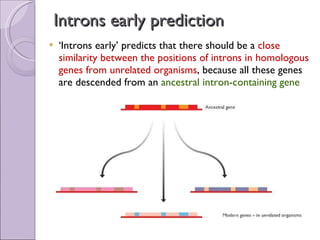
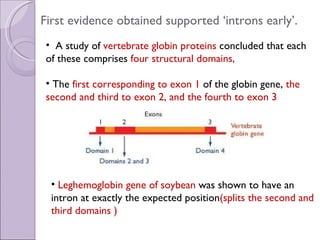
3. Introns may have evolved early in ancestral genes and are gradually being lost from genomes ("introns early" hypothesis) or evolved more recently and are accumulating ("introns late"). Early evidence supported "introns early" with conserved intron positions in homologous genes.