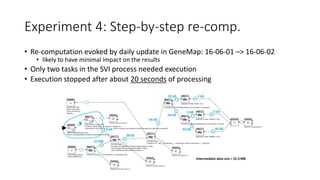
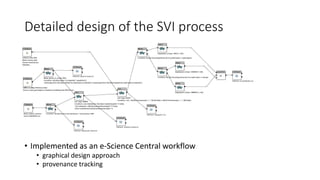
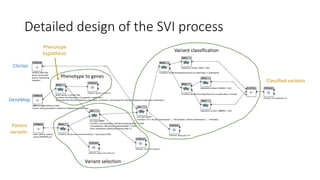
The document discusses the challenges and methodologies for improving efficiency in Next Generation Sequencing (NGS) pipelines, particularly focusing on simple variant identification (SVI). It highlights the need for selective re-computation to minimize costs associated with processing updated genomic data while maintaining accuracy. The experiments demonstrate various approaches to optimize variant classification by assessing changes in input data and refining the re-execution process.





![Experiments: Input data set
Phenotype hypothesis Variant file Variant count File size [MB]
Congenital myasthenic
syndrome
MUN0785 26508 35.5
MUN0789 26726 35.8
MUN0978 26921 35.8
MUN1000 27246 36.3
Parkinsons disease C0011 23940 38.8
C0059 24983 40.4
C0158 24376 39.4
C0176 24280 39.4
Creutzfeldt-Jakob disease A1340 23410 38.0
A1356 24801 40.2
A1362 24271 39.2
A1370 24051 38.9
Frontotemporal dementia -
Amyotrophic lateral sclerosis
B0307 24052 39.0
C0053 23980 38.8
C0171 24387 39.6
D1049 24473 39.5](https://image.slidesharecdn.com/sviunderrecompcontrol-161111164903/85/ReComp-and-the-Variant-Interpretations-Case-Study-11-320.jpg)
![Experiments: Reference data sets
• Different rate of changes:
• GeneMap changes daily
• ClinVar changes monthly
Database Version
Record
count
File size
[MB]
OMIM
GeneMap
2016-03-08 13053 2.2
2016-04-28 15871 2.7
2016-06-01 15897 2.7
2016-06-02 15897 2.7
2016-06-07 15910 2.7
NCBI ClinVar 2015-02 281023 96.7
2016-02 285041 96.6
2016-05 290815 96.1](https://image.slidesharecdn.com/sviunderrecompcontrol-161111164903/85/ReComp-and-the-Variant-Interpretations-Case-Study-12-320.jpg)

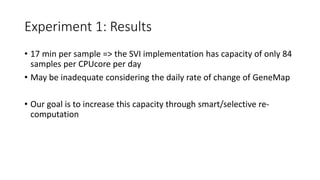
![Experiment 1: Results
• Running the SVI workflow on one patient sample takes about 17
minutes
• executed on a single-core VM
• may be optimised –> optimisation out-of-scope at the moment
• Runtime is consistent across different phenotypes
• Changes of the GeneMap and ClinVar version have negligible impact
on the execution time, e.g.:
Run time [mm:ss]
GeneMap version 2016-03-08 2016-04-28 2016-06-07
μ ± σ 17:05 ± 22 17:09 ± 15 17:10 ± 17](https://image.slidesharecdn.com/sviunderrecompcontrol-161111164903/85/ReComp-and-the-Variant-Interpretations-Case-Study-14-320.jpg)

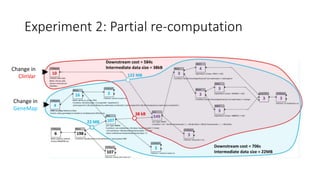
![Experiment 2: Results
• Running the part of SVI directly involved in
processing updated data can save some
runtime
• Savings depend on:
• the structure of the process
• the point where the changed data are used
• Savings involve the cost of retaining interim
data required in partial re-execution
• the size of the data depends on the
phenotype hypothesis and type of change
• the size is in range of 20–22 MB for GeneMap
changes and 2–334 kB for ClinVar changes
Run time
[mm:ss]
Savings Run time
[mm:ss]
Savings
GeneMap
version
2016-04-28 2016-06-07
μ ± σ 11:51 ± 16 31% 11:50 ± 20 31%
ClinVar
version
2016-02 2016-05
μ ± σ 9:51 ± 14 43% 9:50 ± 15 42%](https://image.slidesharecdn.com/sviunderrecompcontrol-161111164903/85/ReComp-and-the-Variant-Interpretations-Case-Study-18-320.jpg)

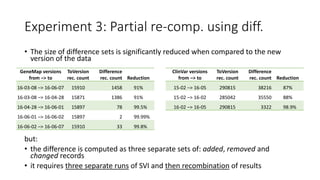
![Experiment 3: Results
• Running the part of SVI directly involved in
processing updated data can save some
runtime
• Running the part of SVI on each difference set
also saves some runtime
• Yet, the total cost of three separate re-
executions may exceed the savings
• Concluding, this approach has a few weak
points:
• running the process on diff. sets is not always
possible
• running the process using diff. sets requires
output recombination
• total runtime may sometimes exceed the
runtime of a regular update
Run time [mm:ss]
Added Removed Changed Total
GeneMap
change
11:30 ± 5 11:27 ± 11 11:36 ± 8 34:34 ± 16
ClinVar
change
2:29 ± 9 0:37 ± 7 0:44 ± 7 3:50 ± 22](https://image.slidesharecdn.com/sviunderrecompcontrol-161111164903/85/ReComp-and-the-Variant-Interpretations-Case-Study-21-320.jpg)