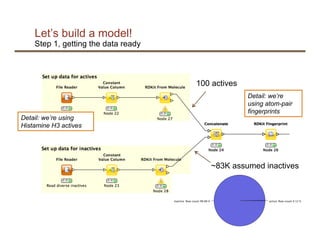
This document discusses using machine learning and the KNIME platform to build predictive models for problems in the life sciences using molecular data. It provides an example of building a random forest model to predict biological activity of molecules using molecular fingerprints as features. The model achieves high accuracy but predicts inactivity for almost all molecules due to class imbalance in the data. To address this, the document suggests adjusting the decision boundary of the model by setting it at the point on the ROC curve that retrieves most actives without including too many inactives. In summary, it presents an example of applying machine learning to predict biological activity from molecular data and discusses techniques for handling class imbalance.