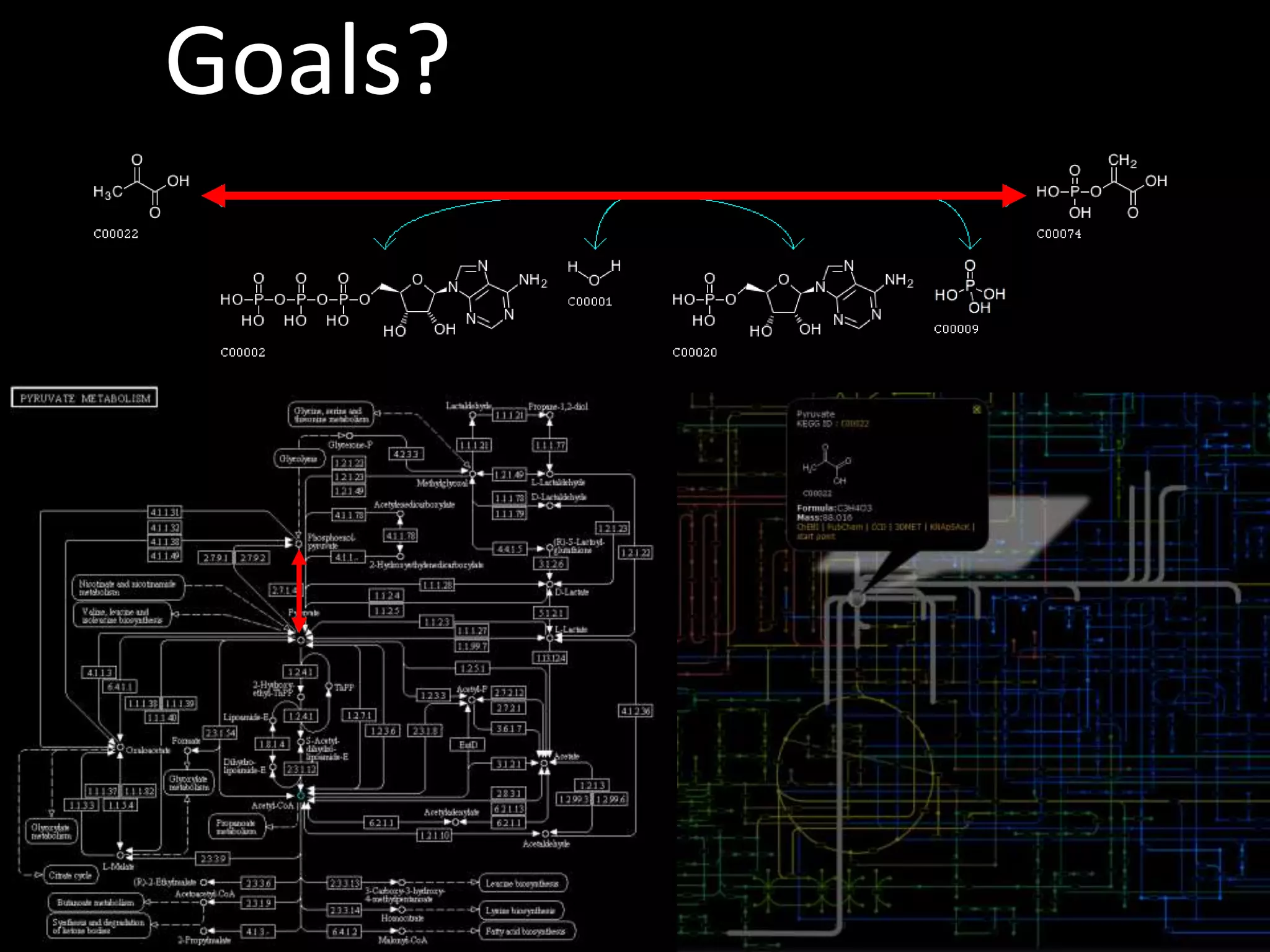
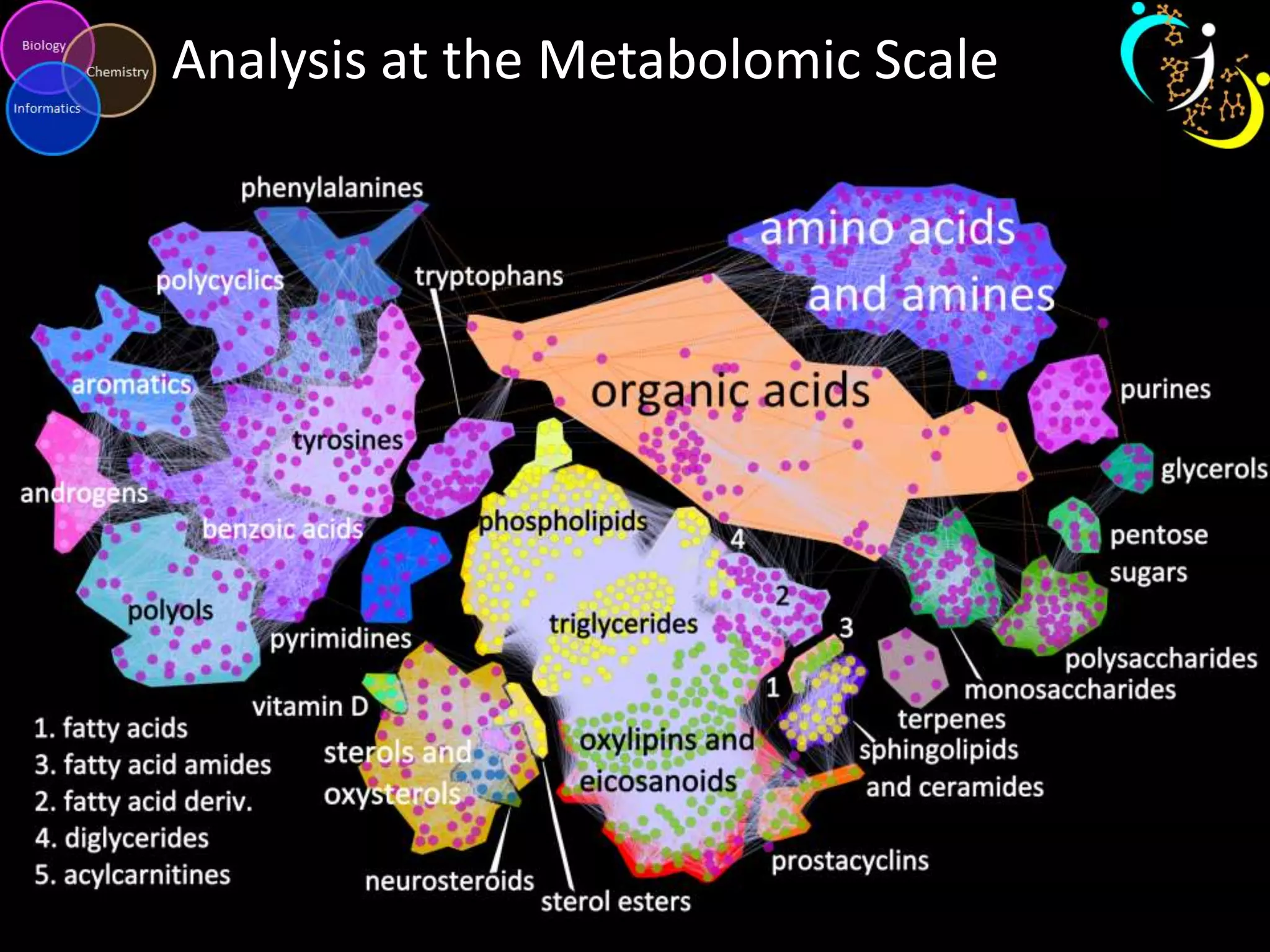
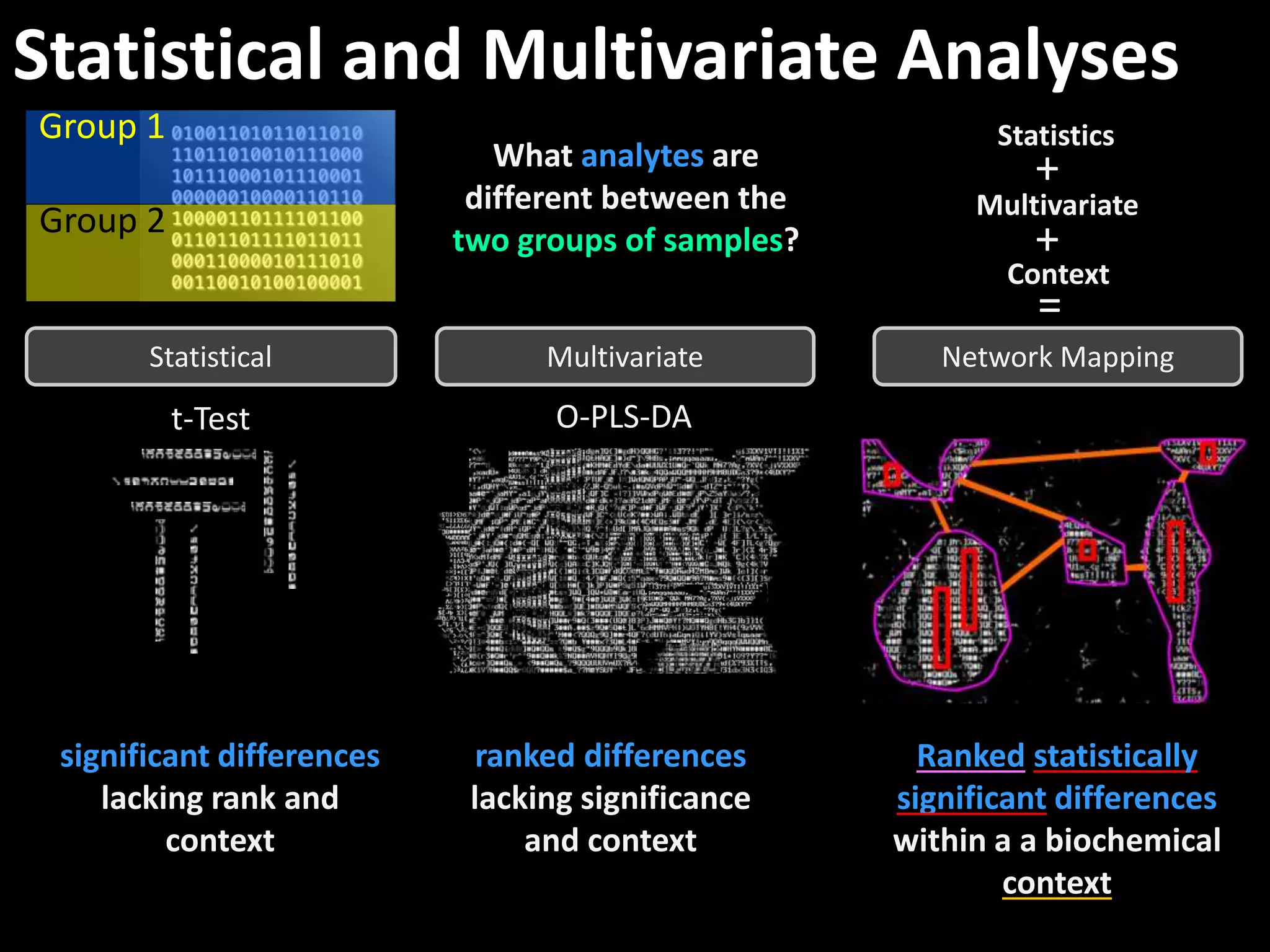
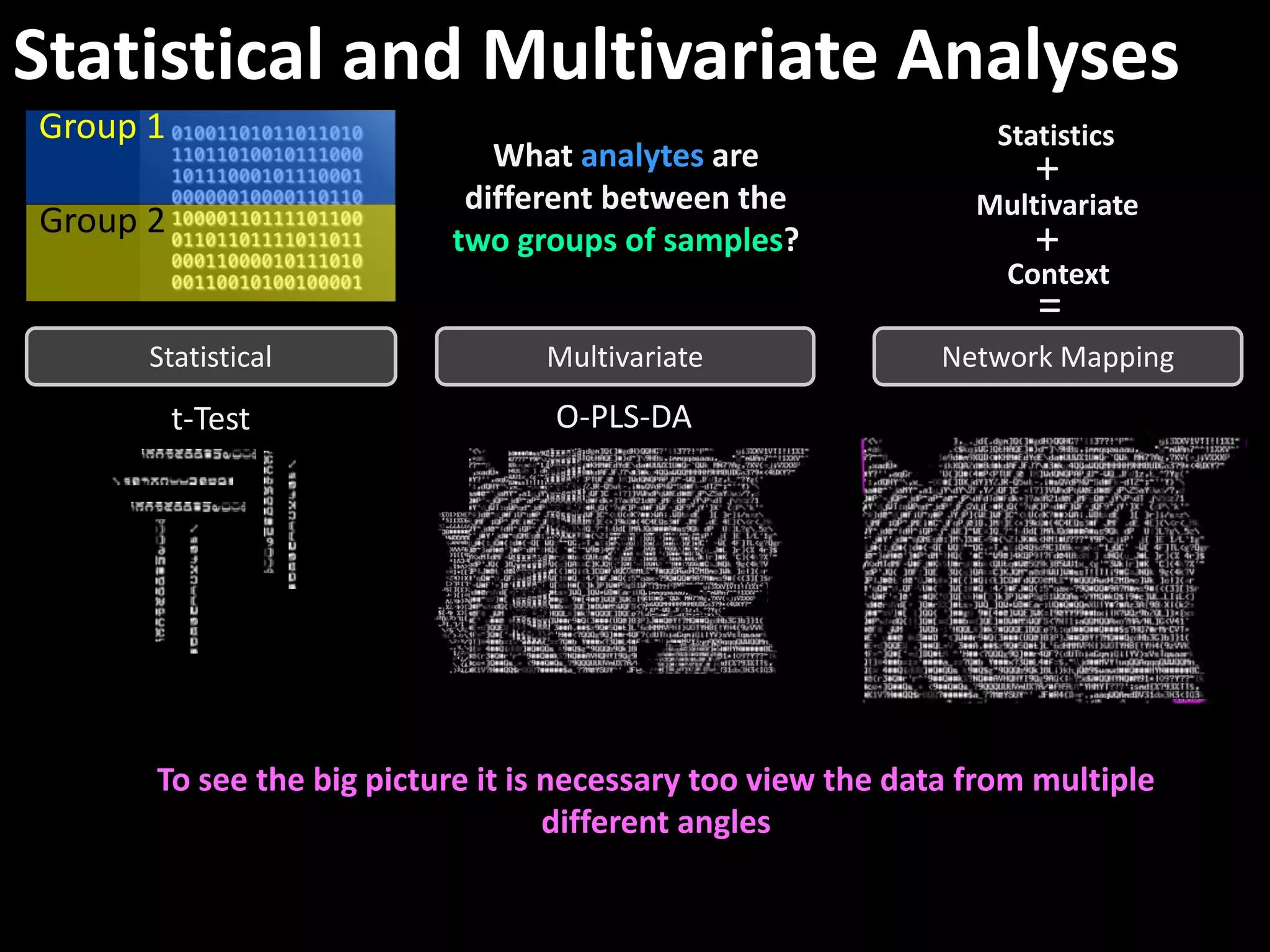
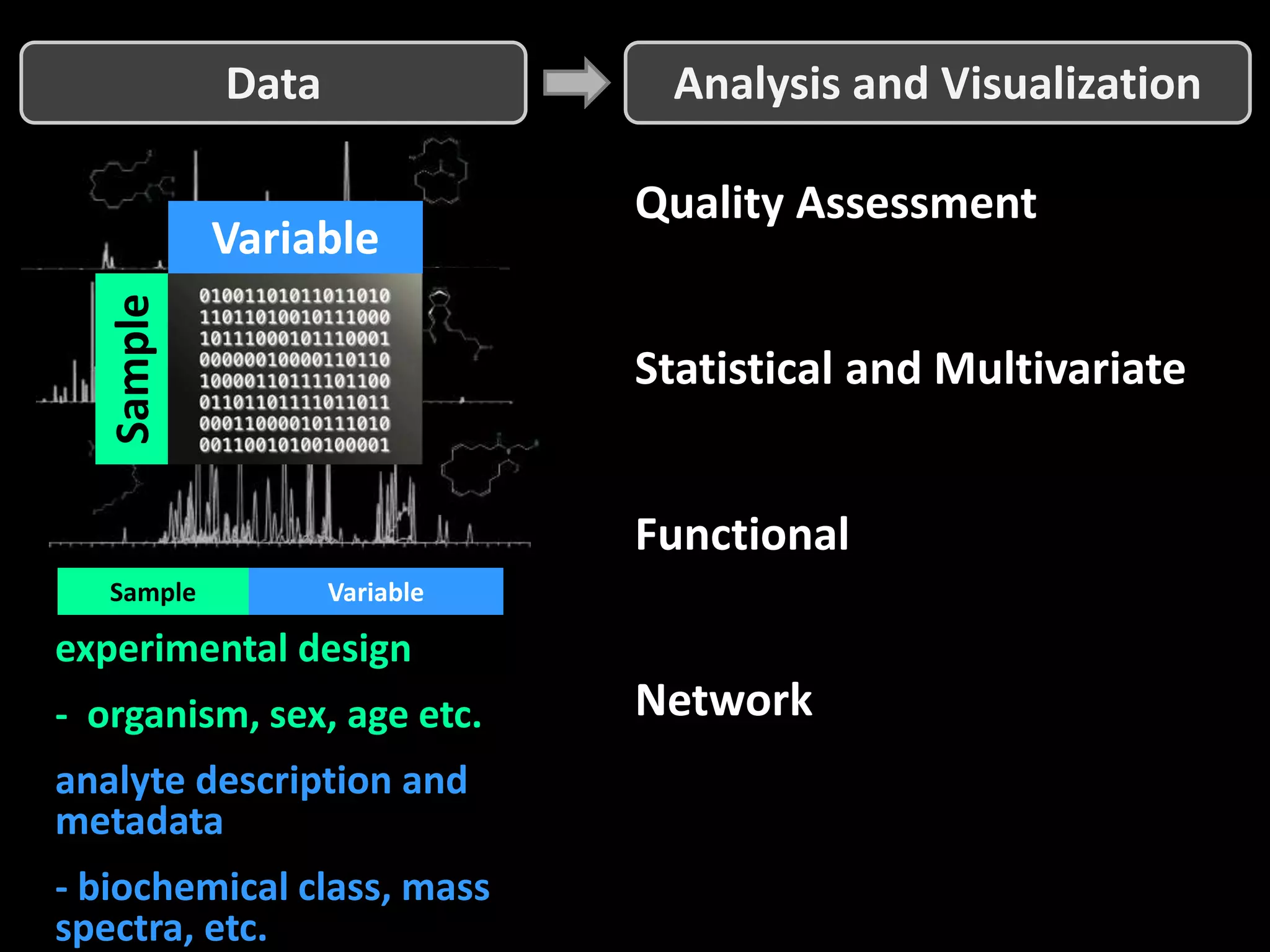
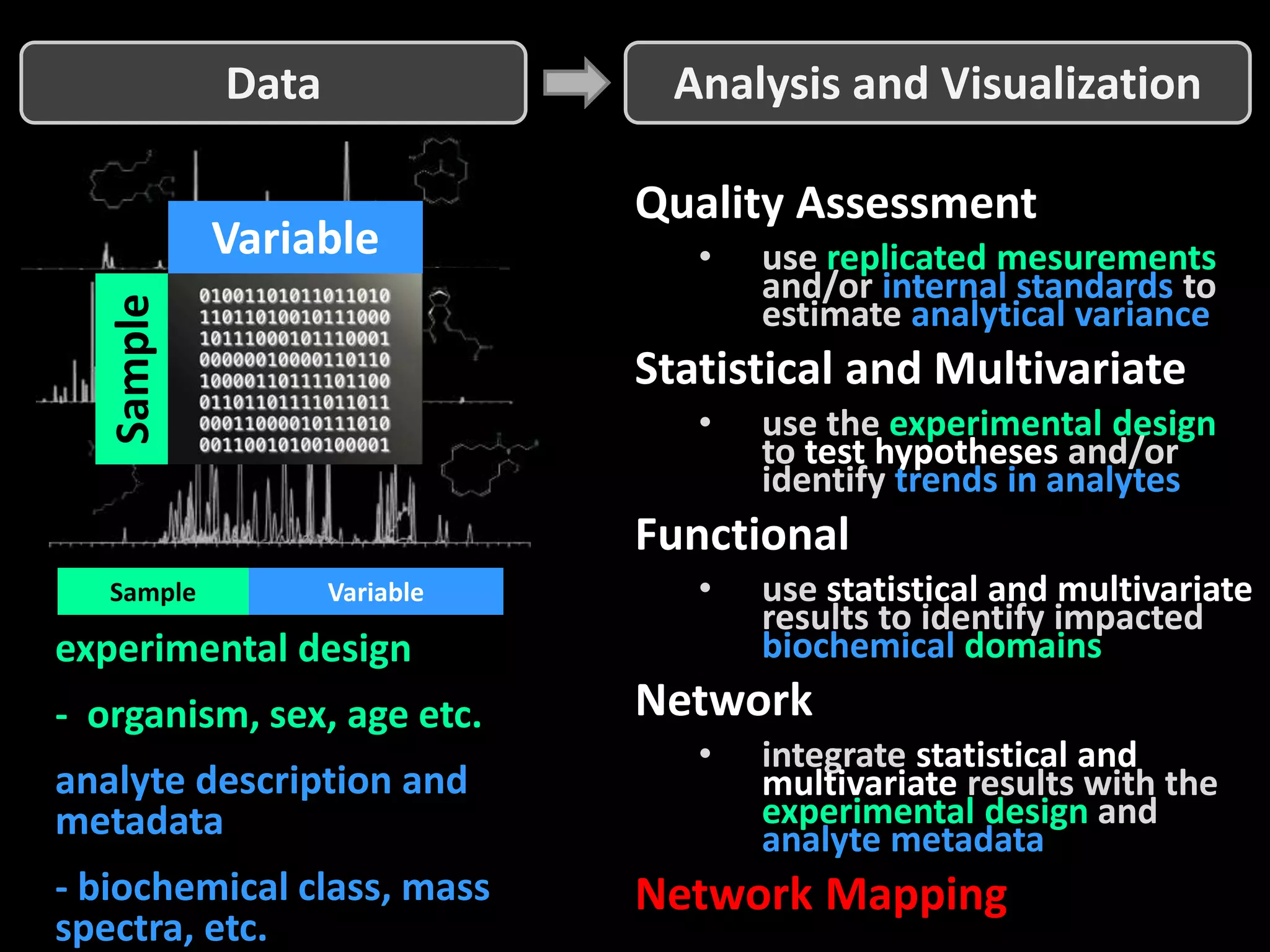
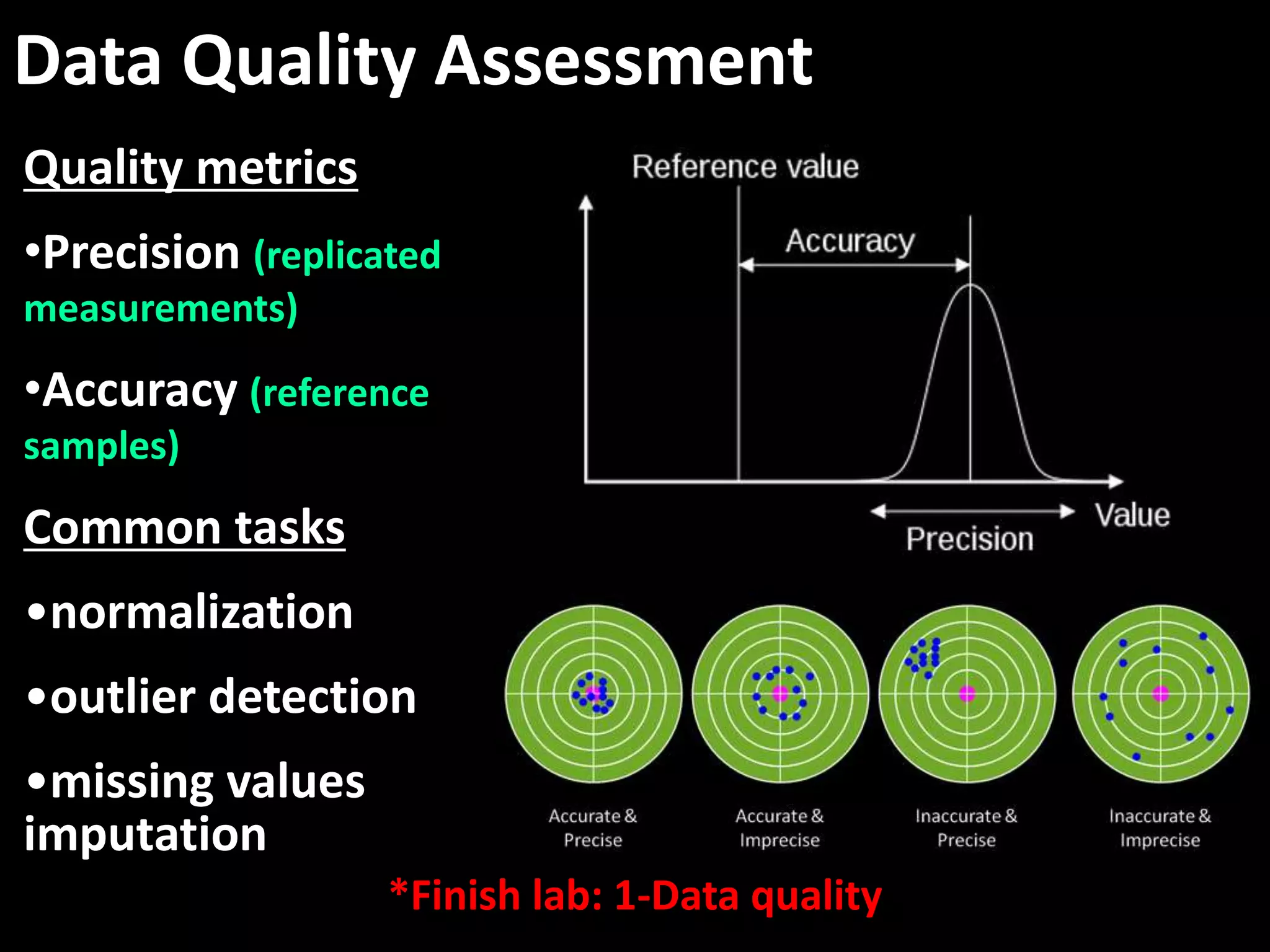
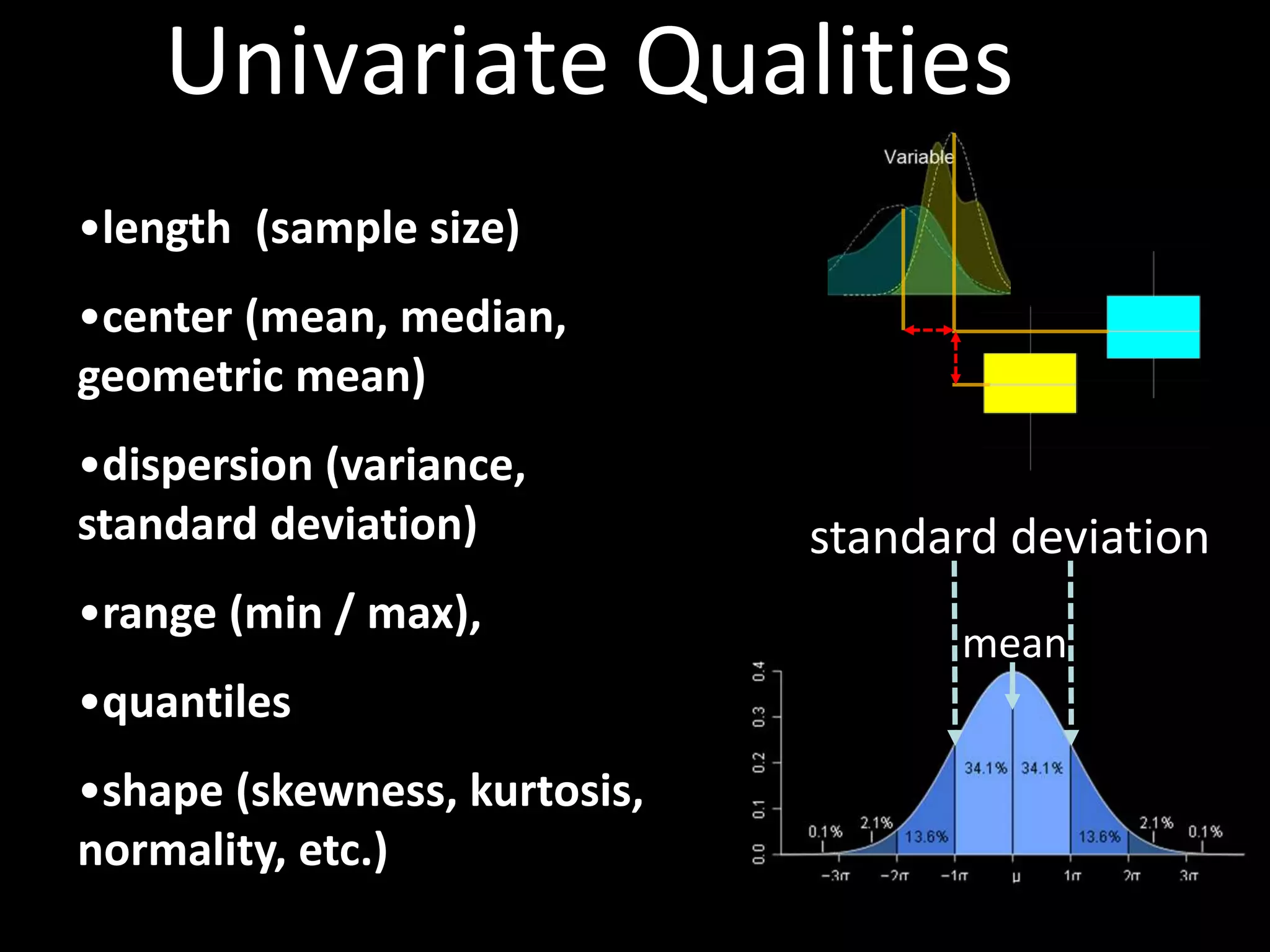
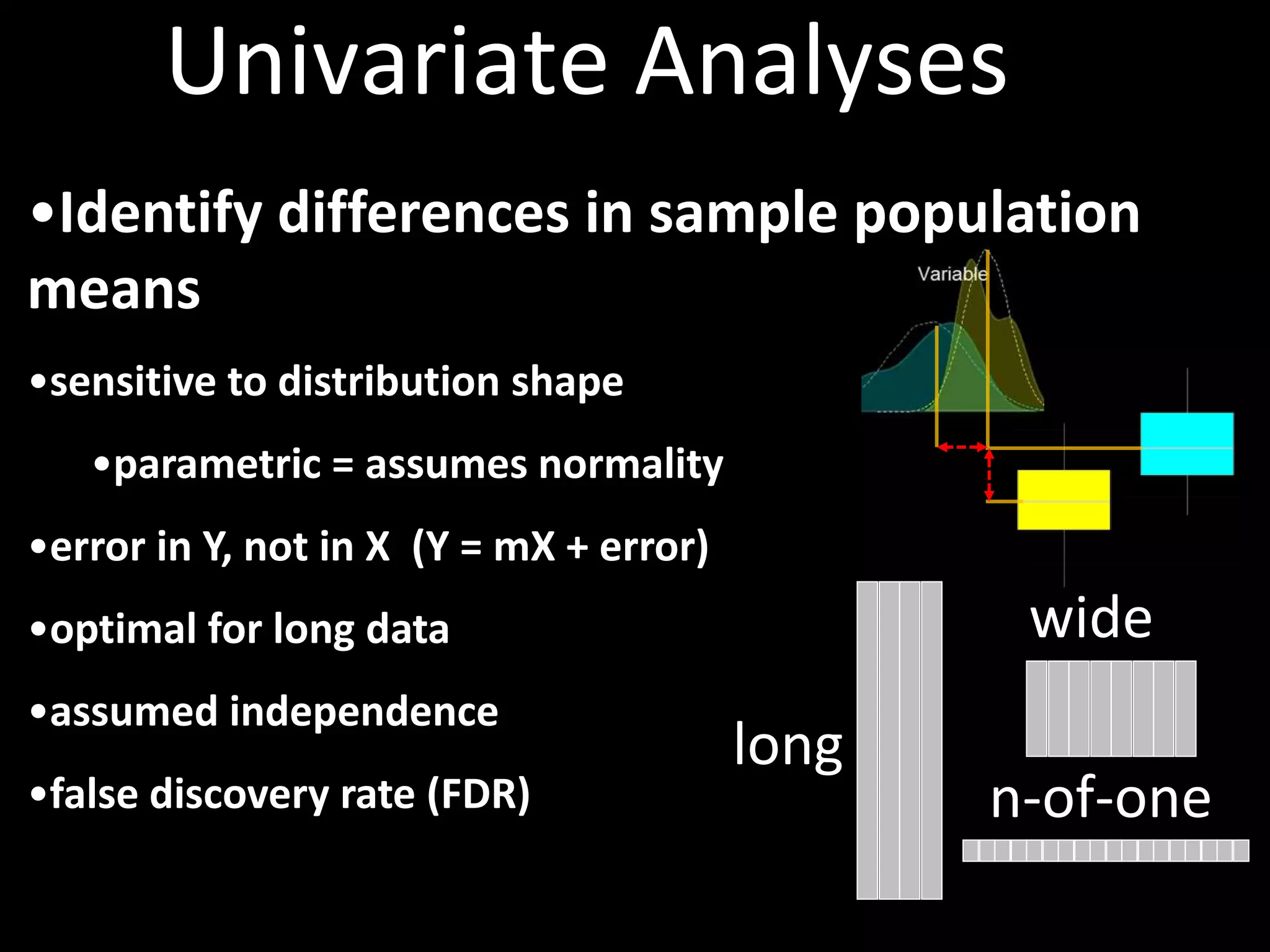
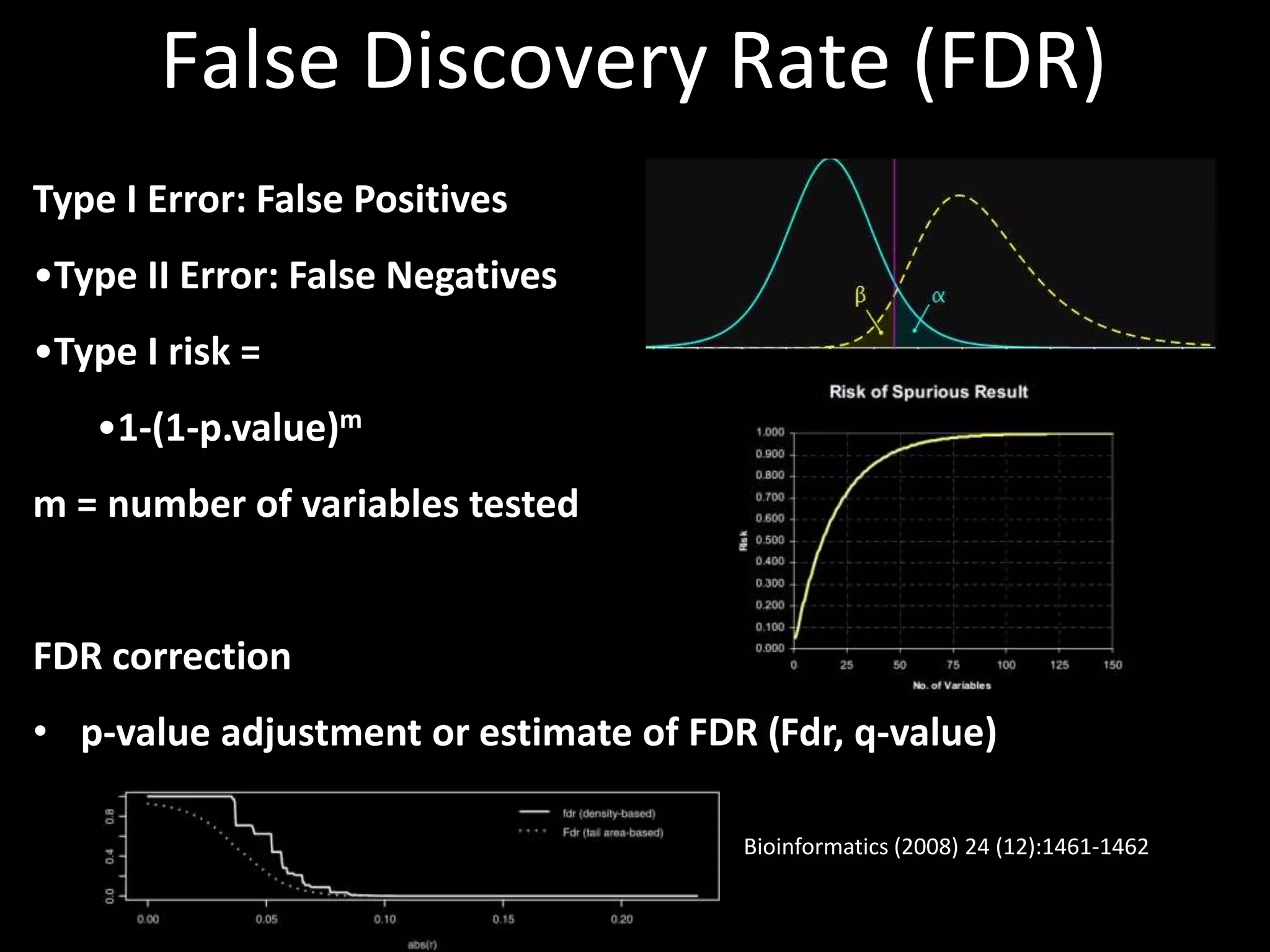
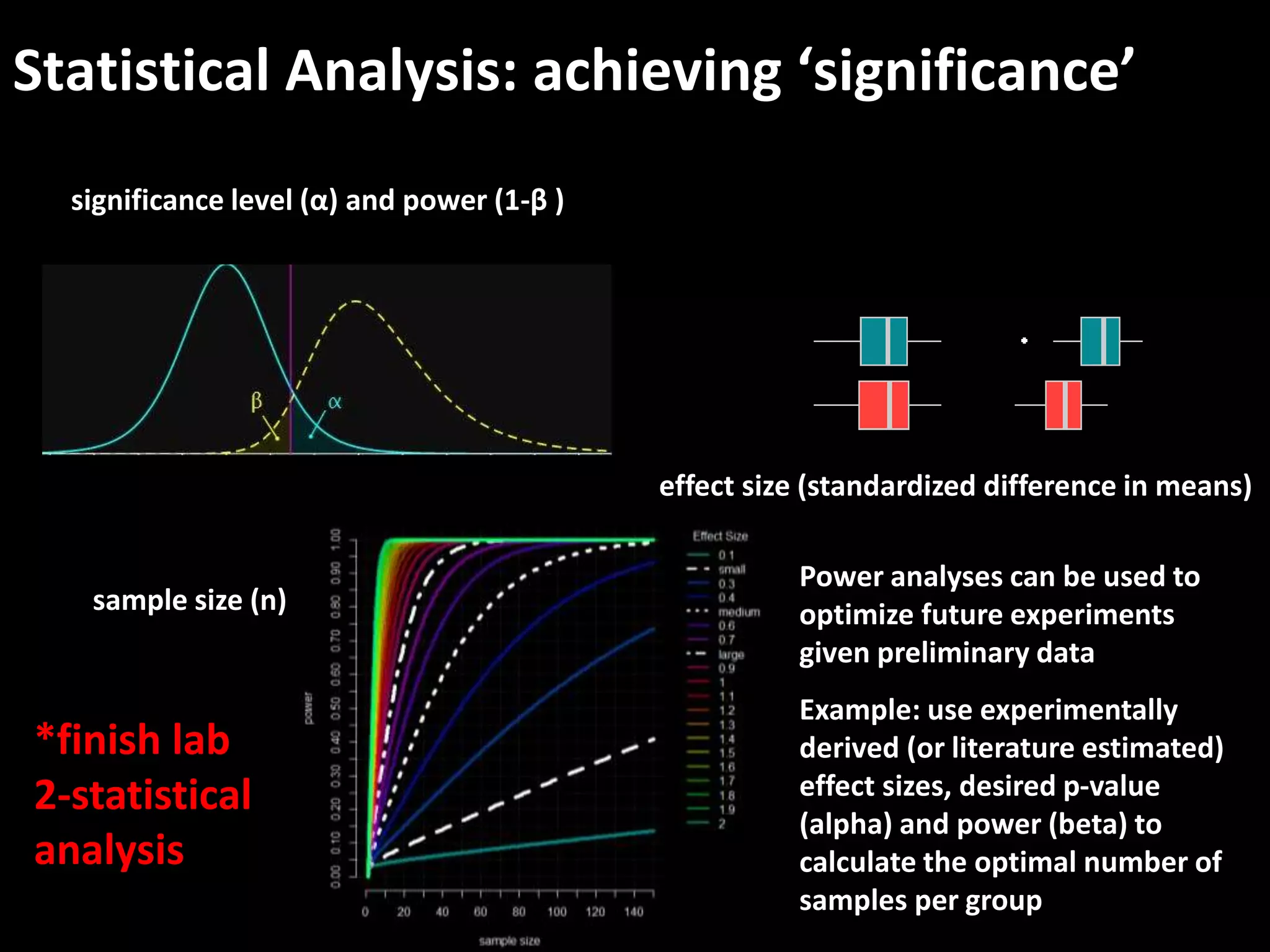
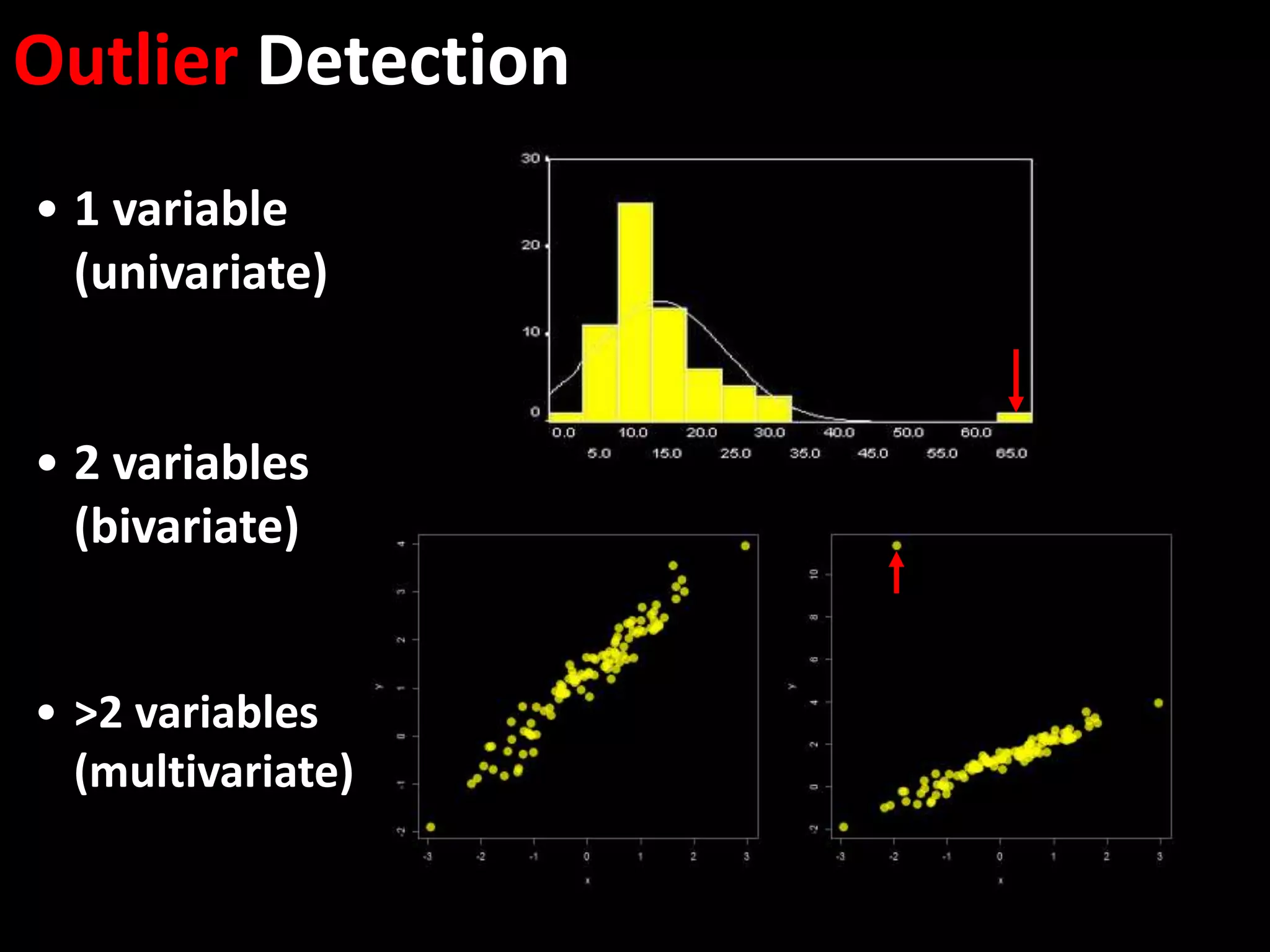
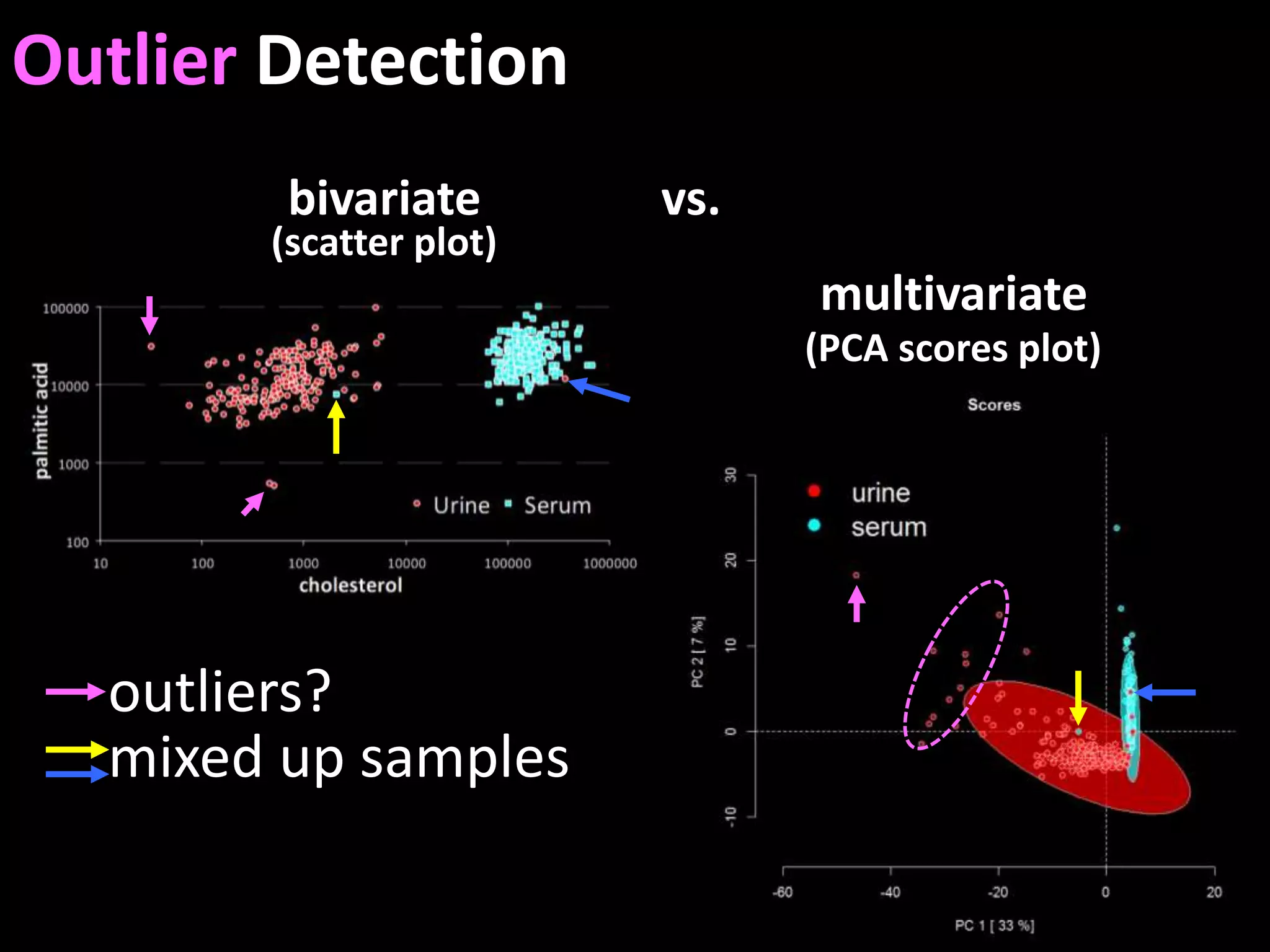
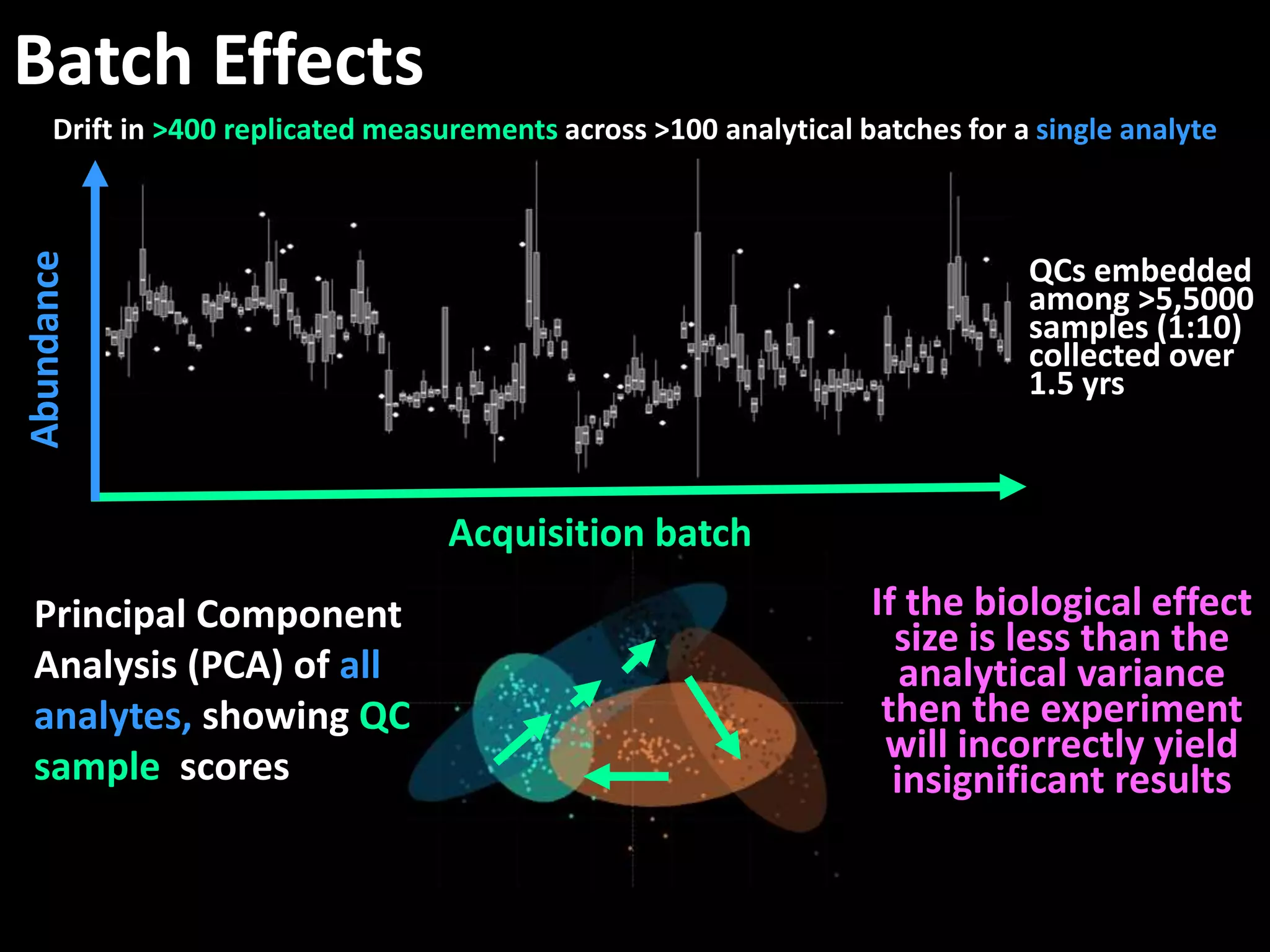
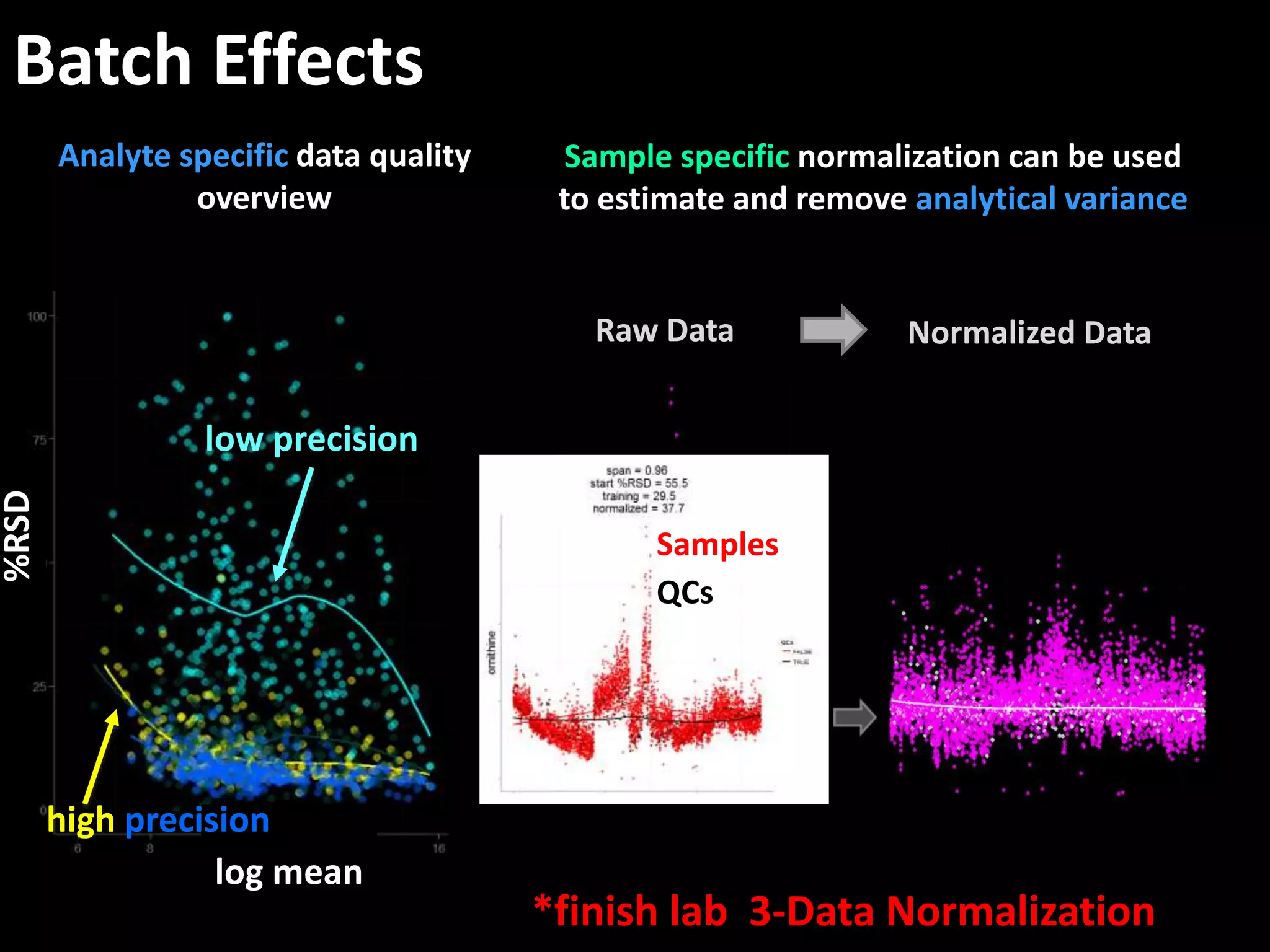
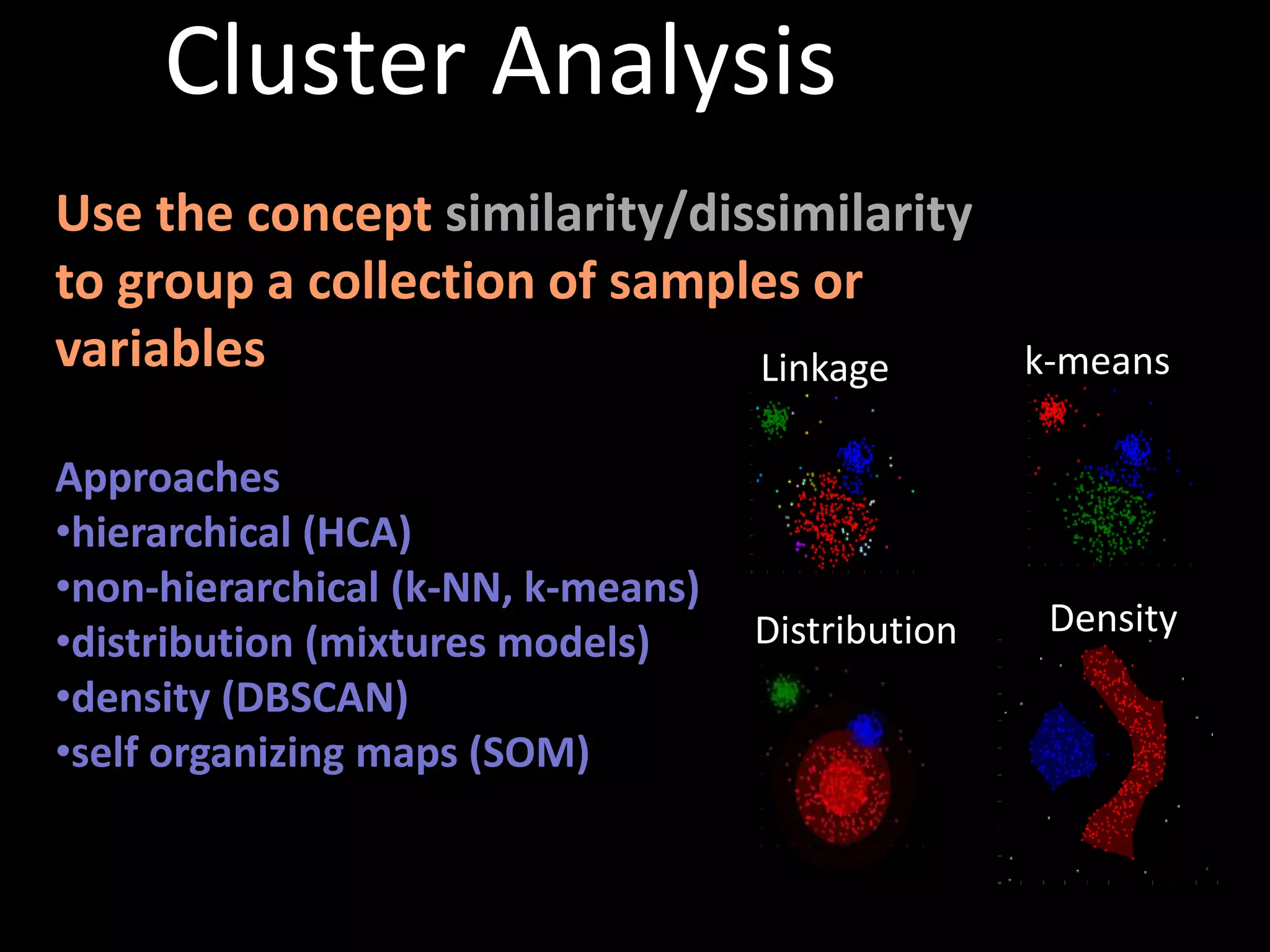
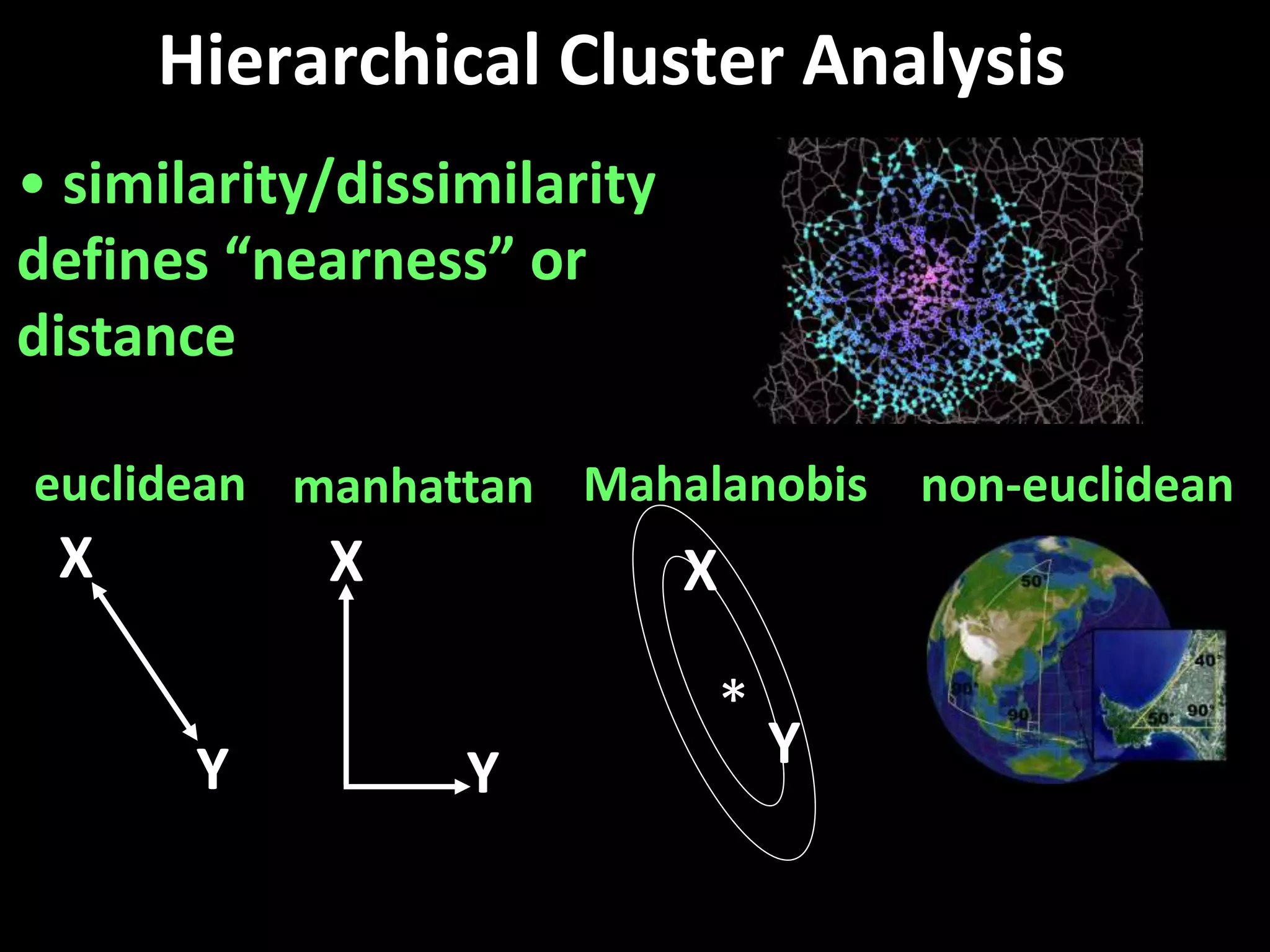
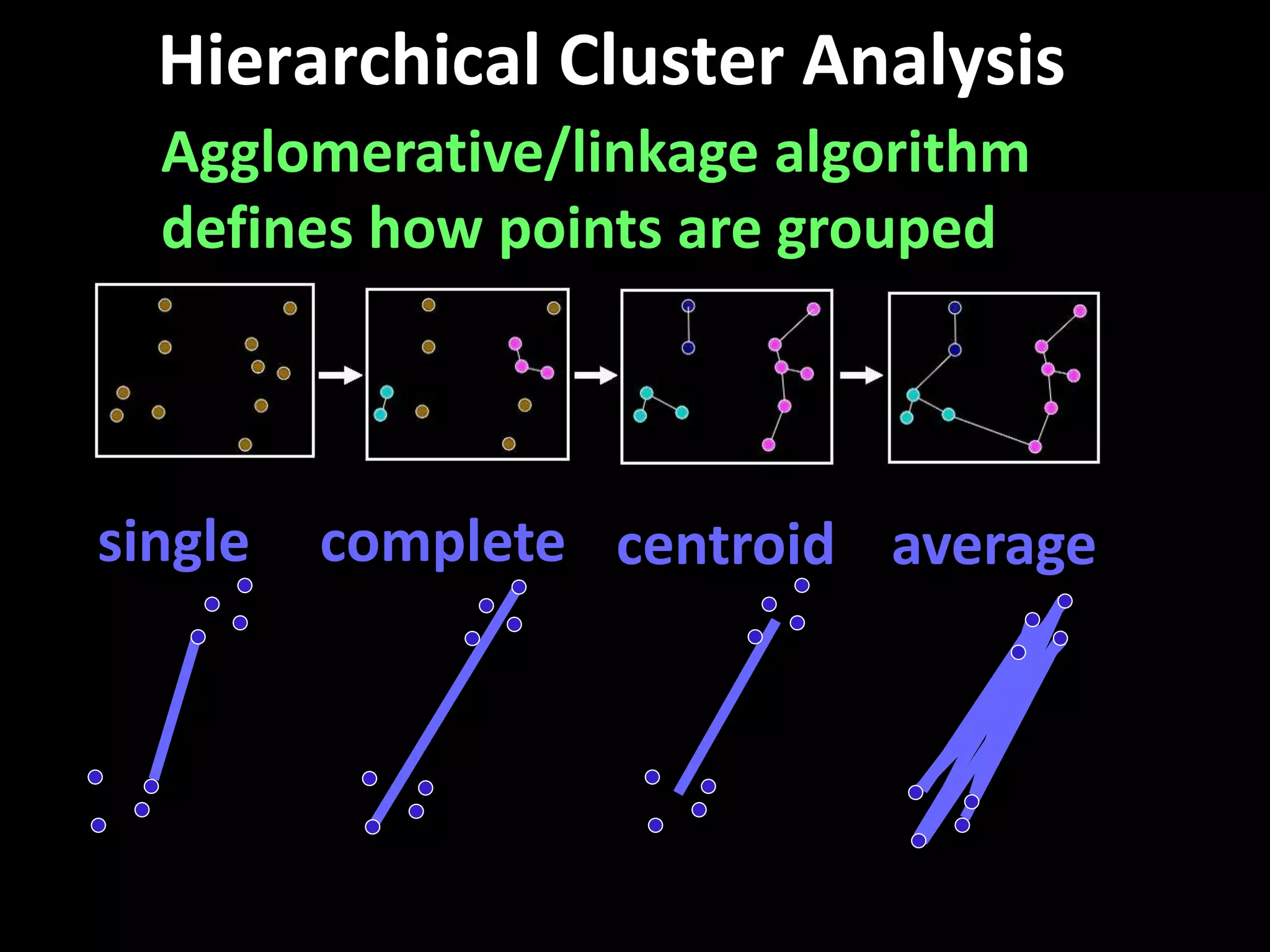
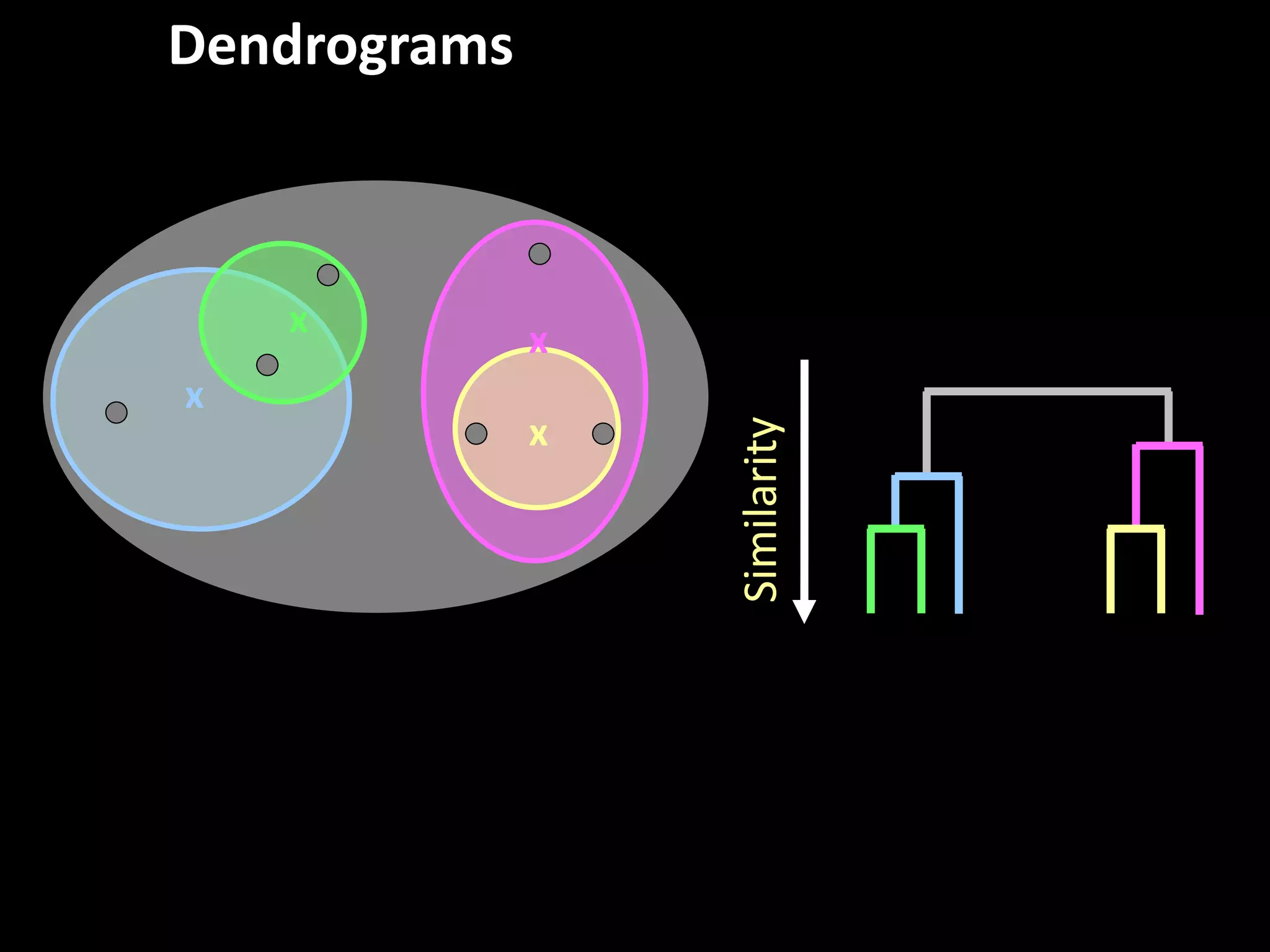
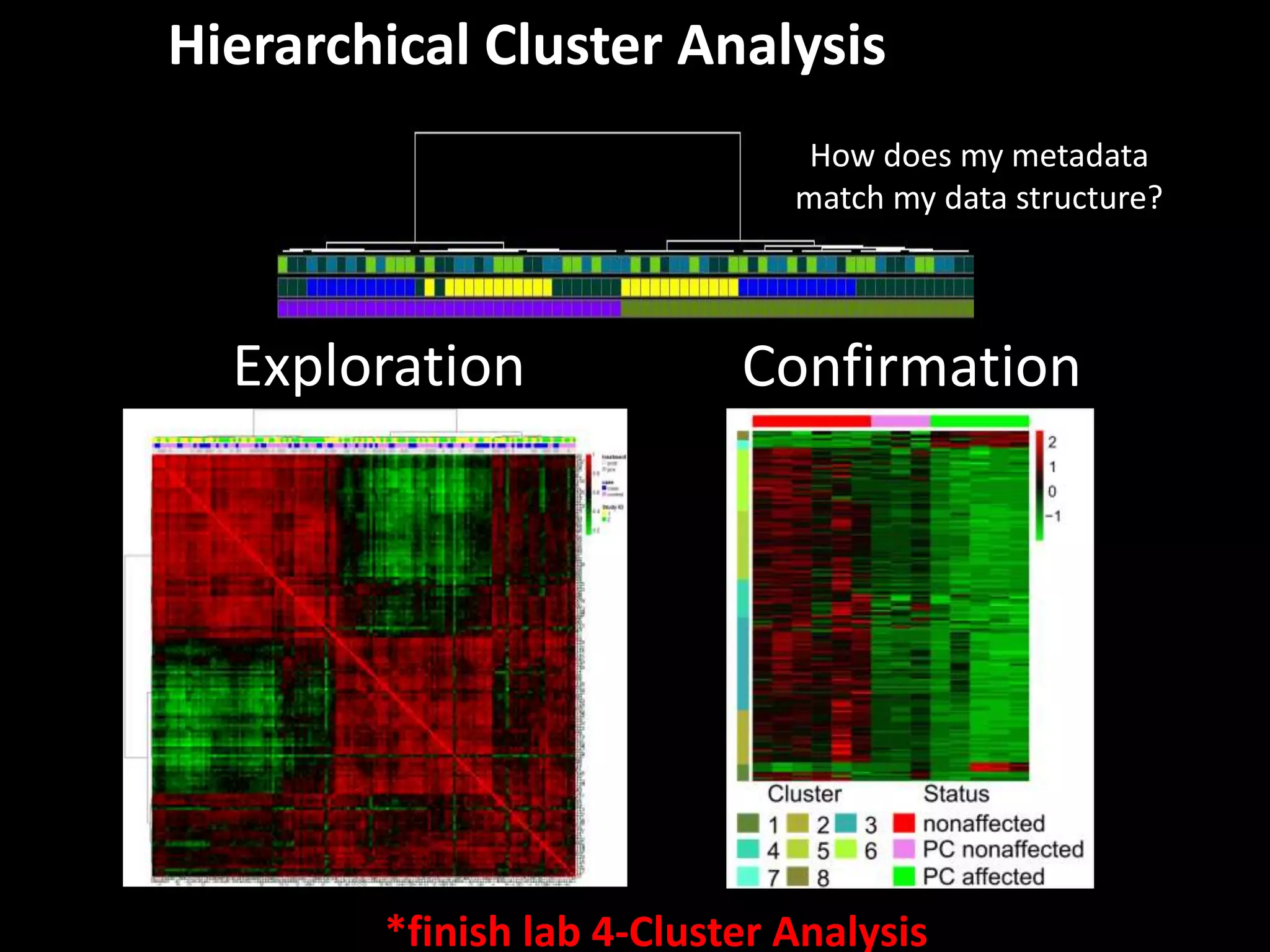
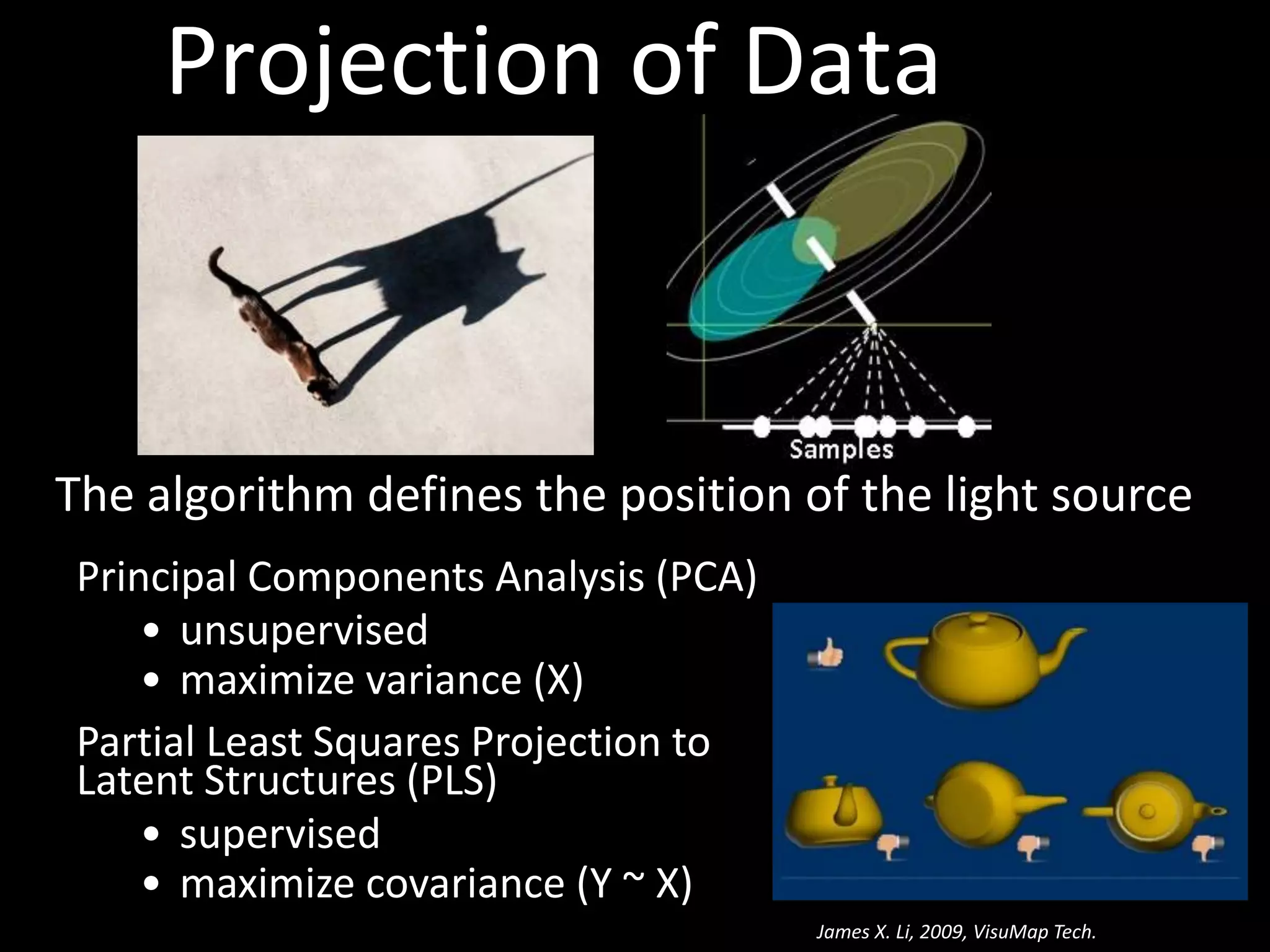
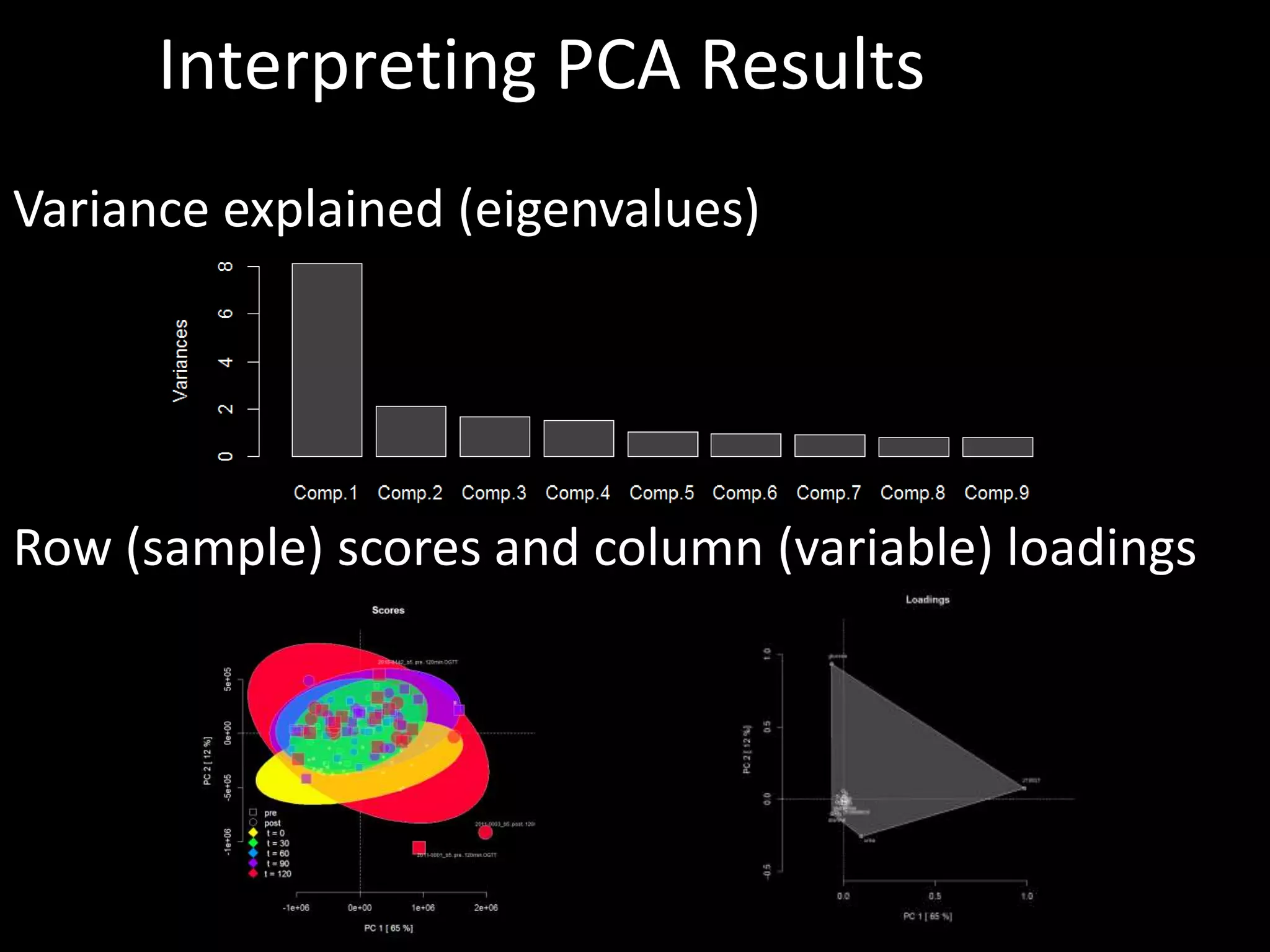
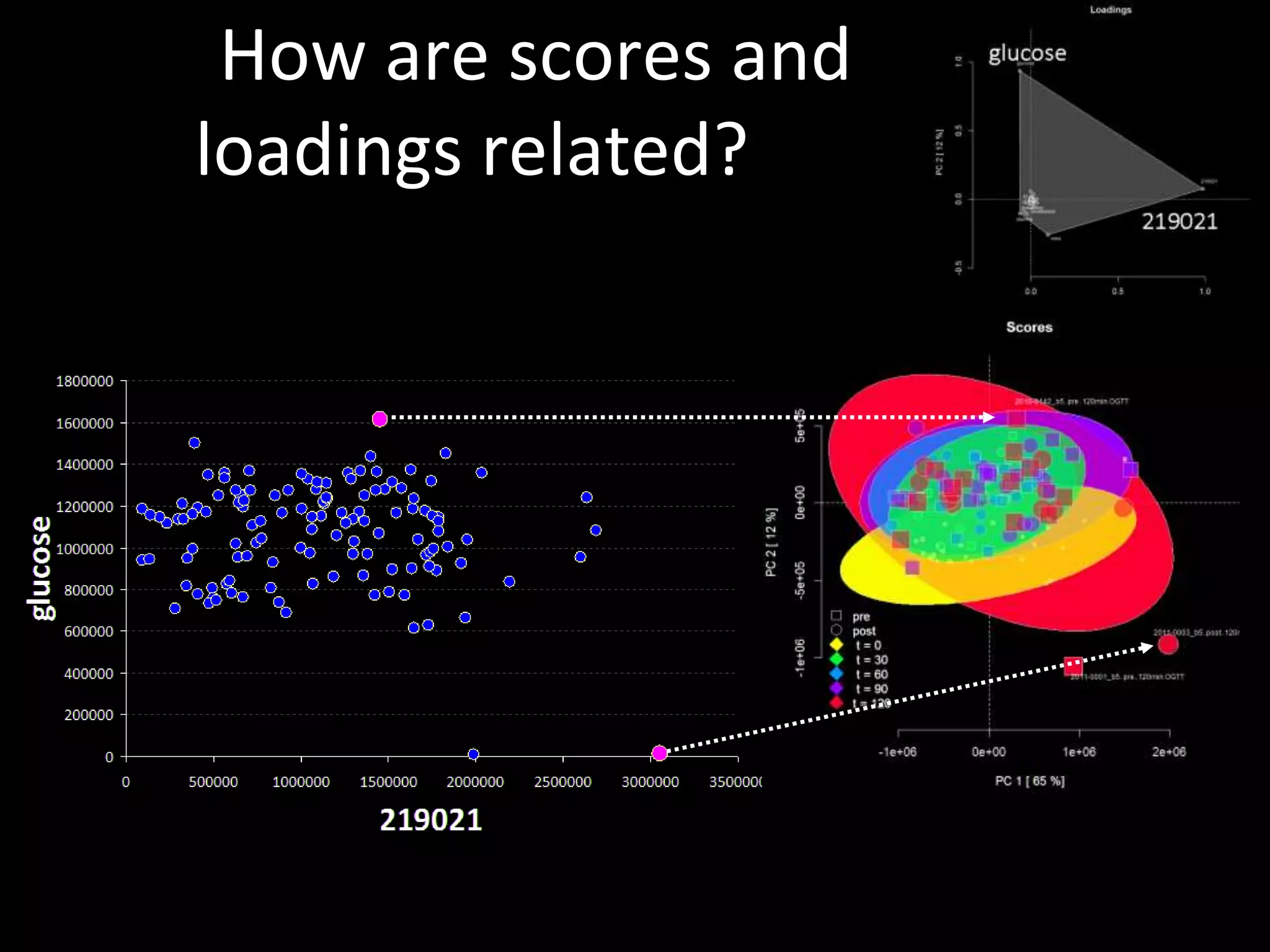
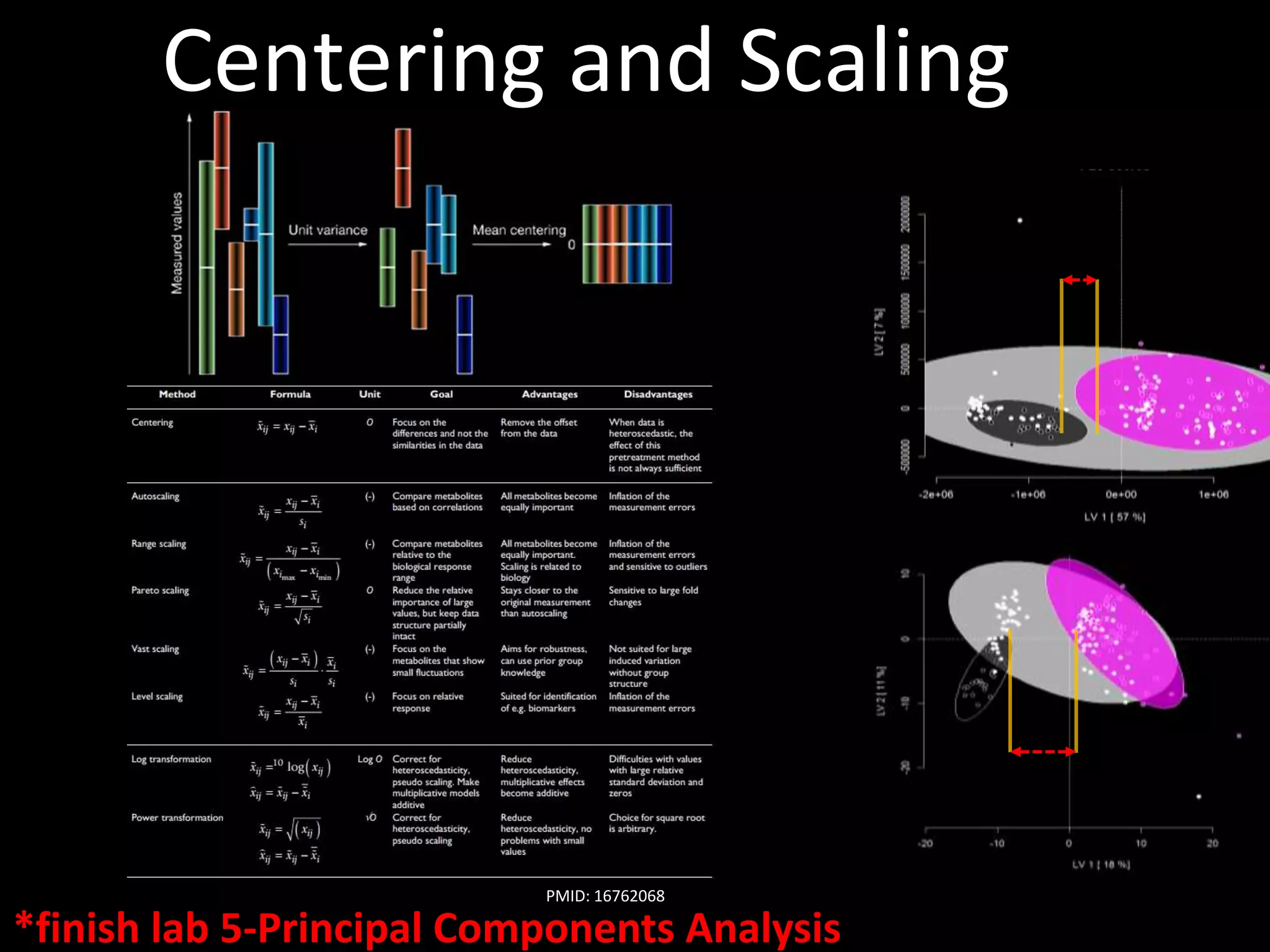
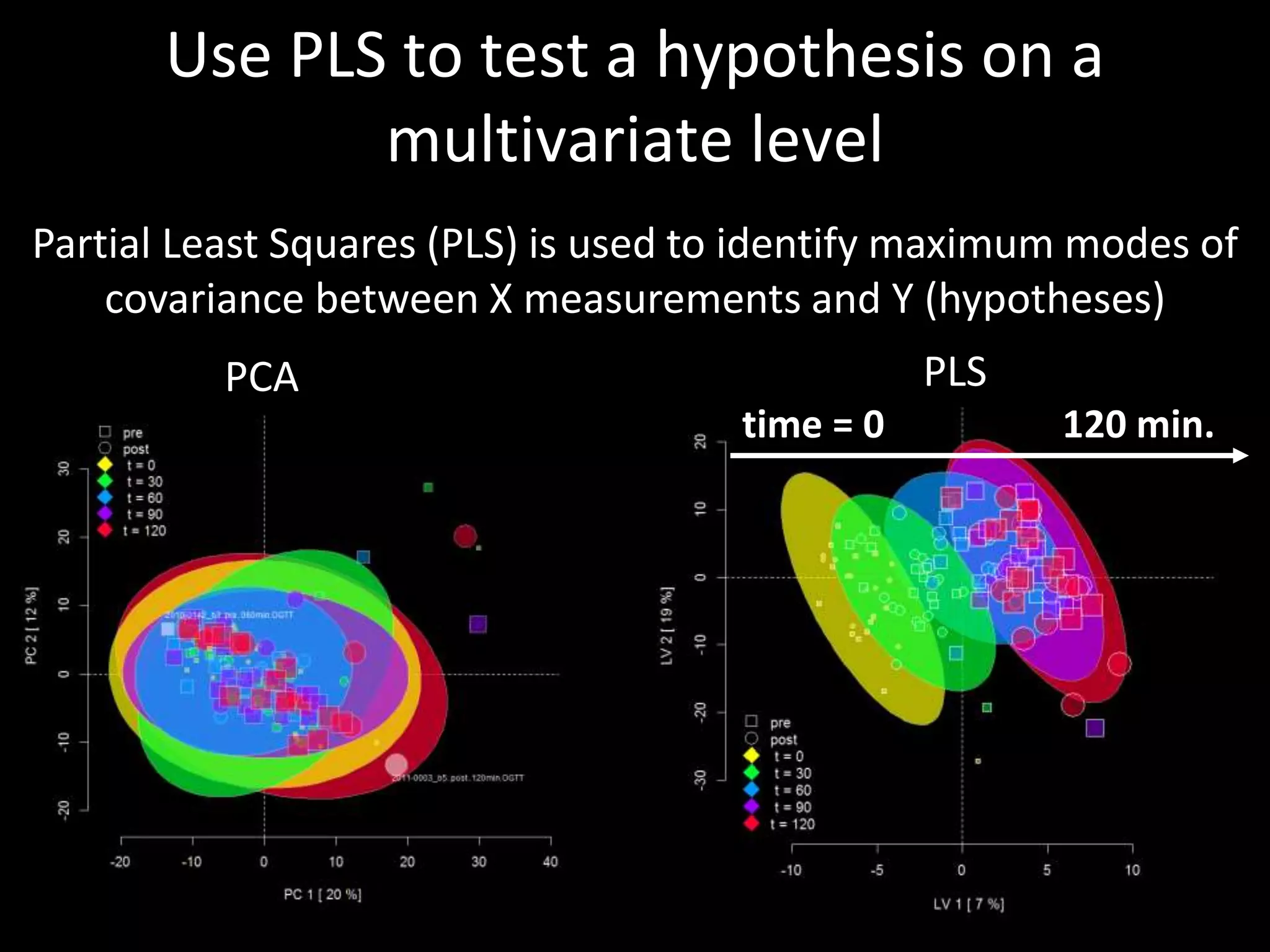
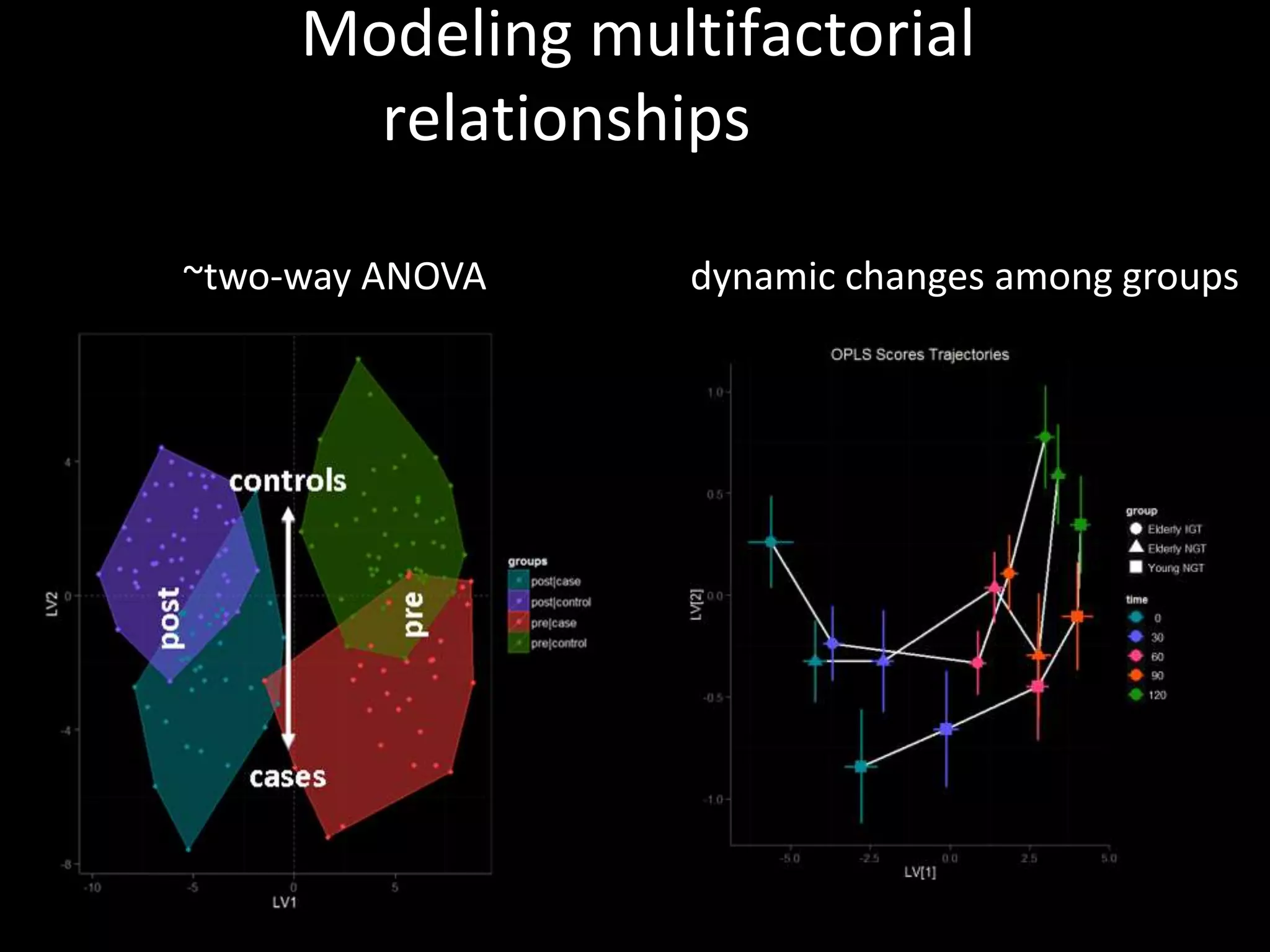
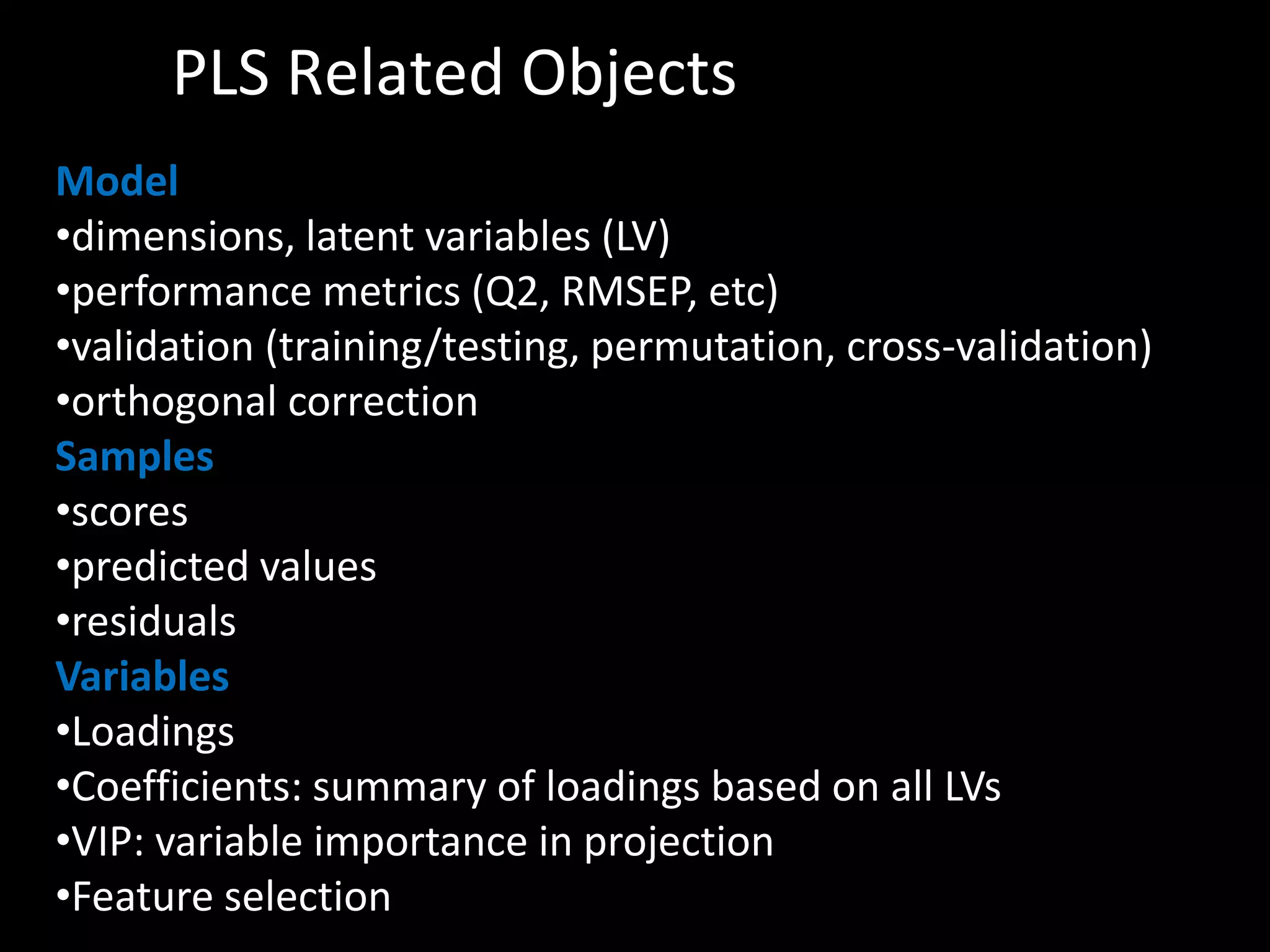
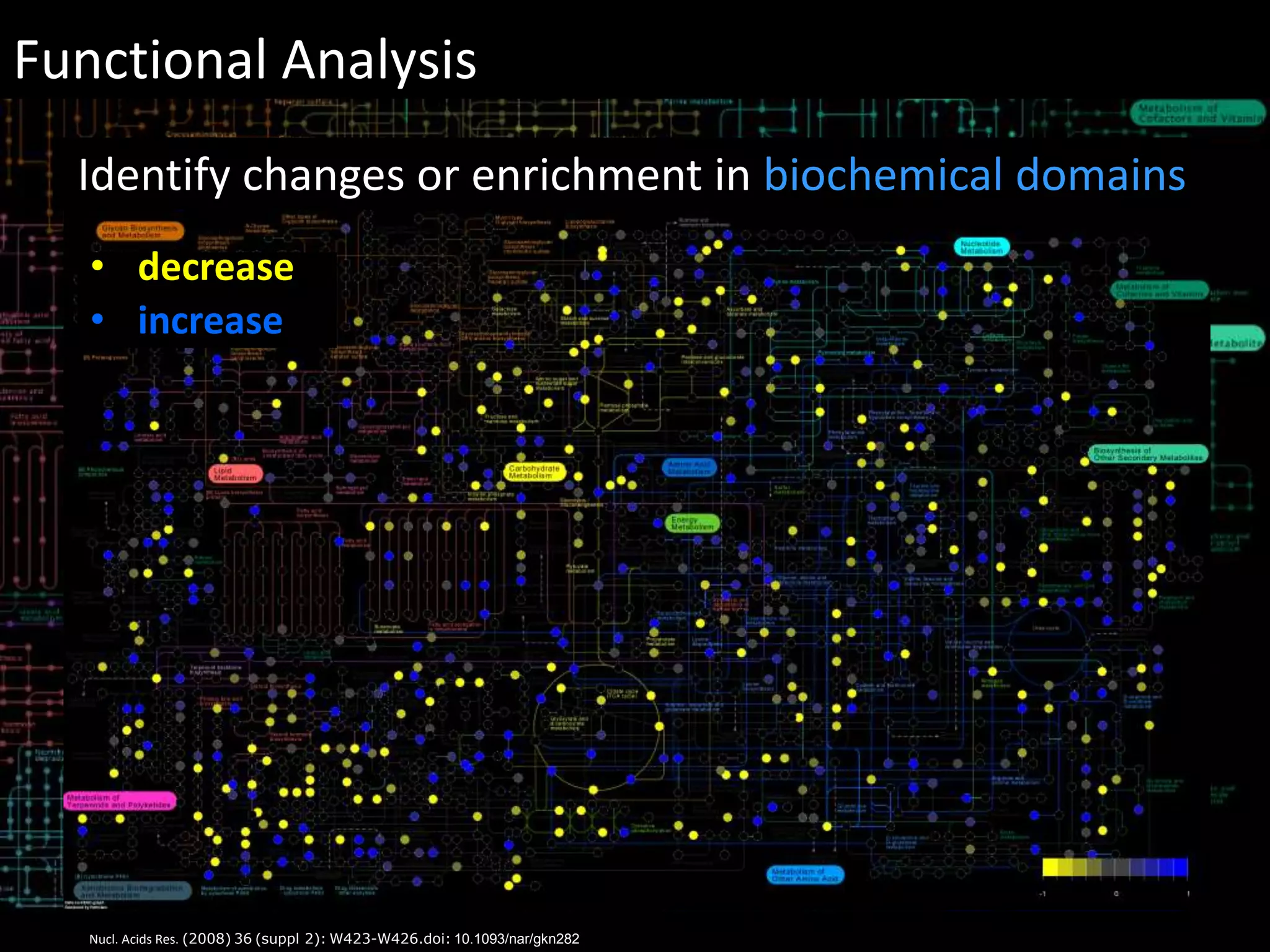
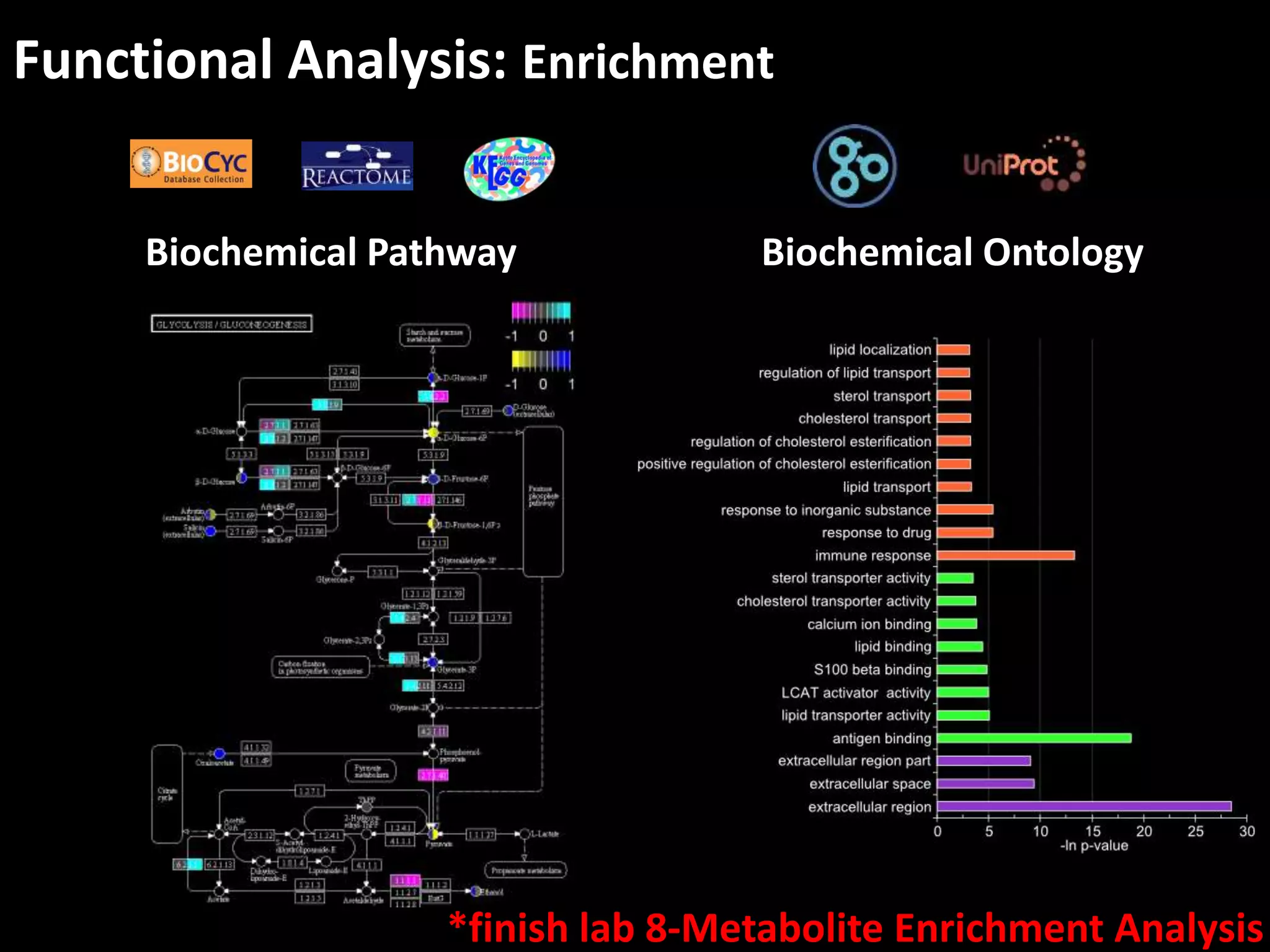
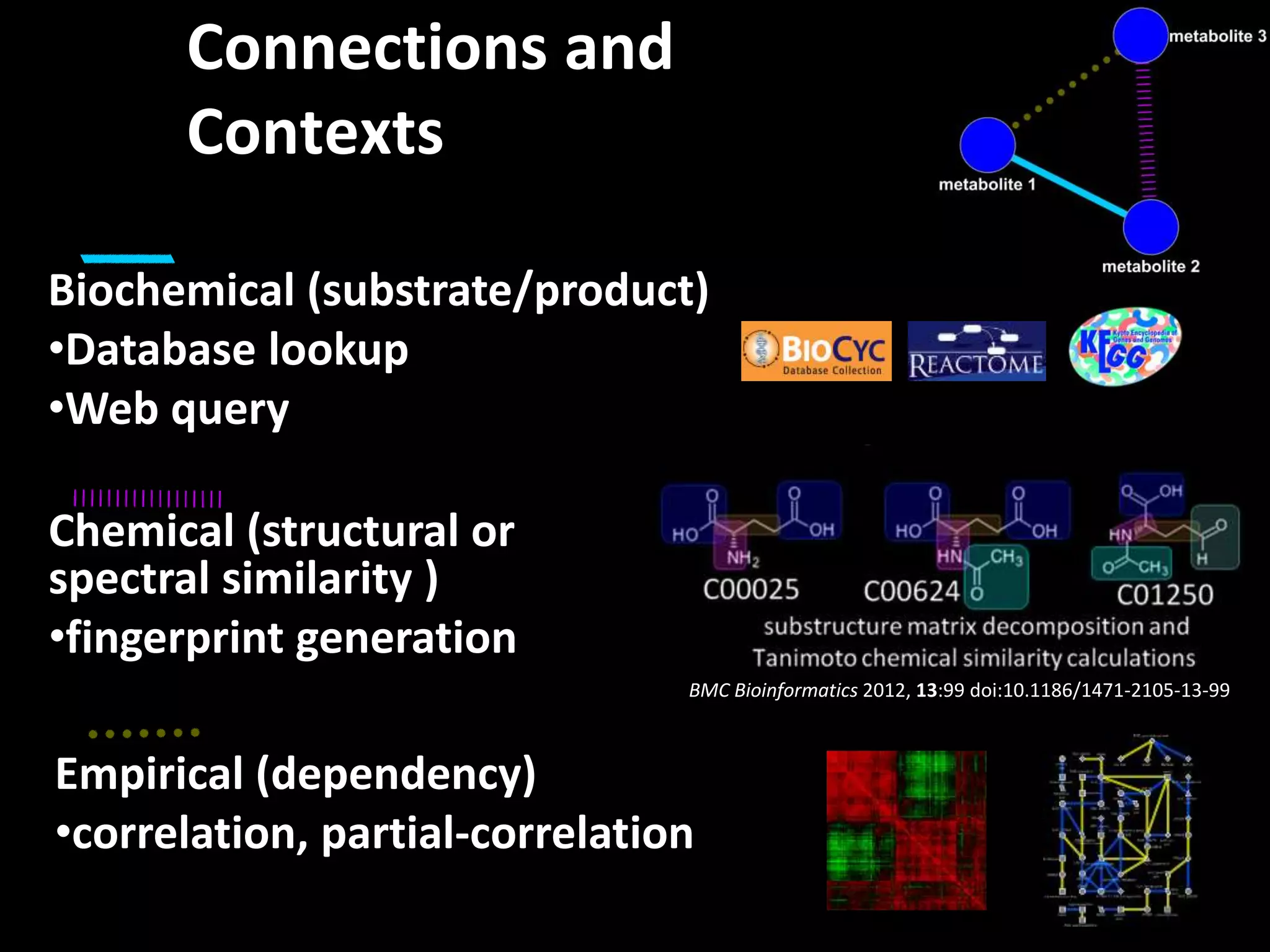
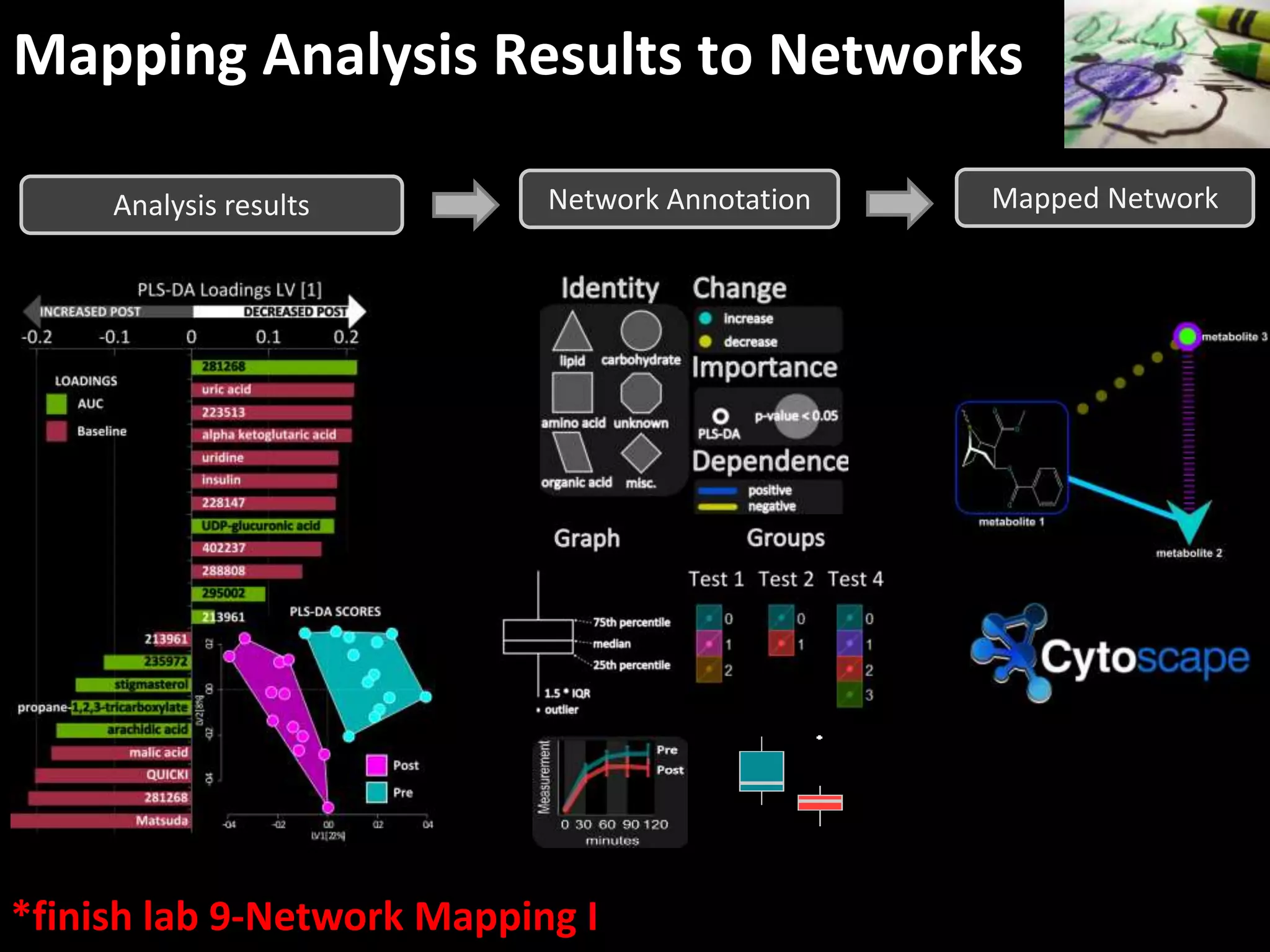
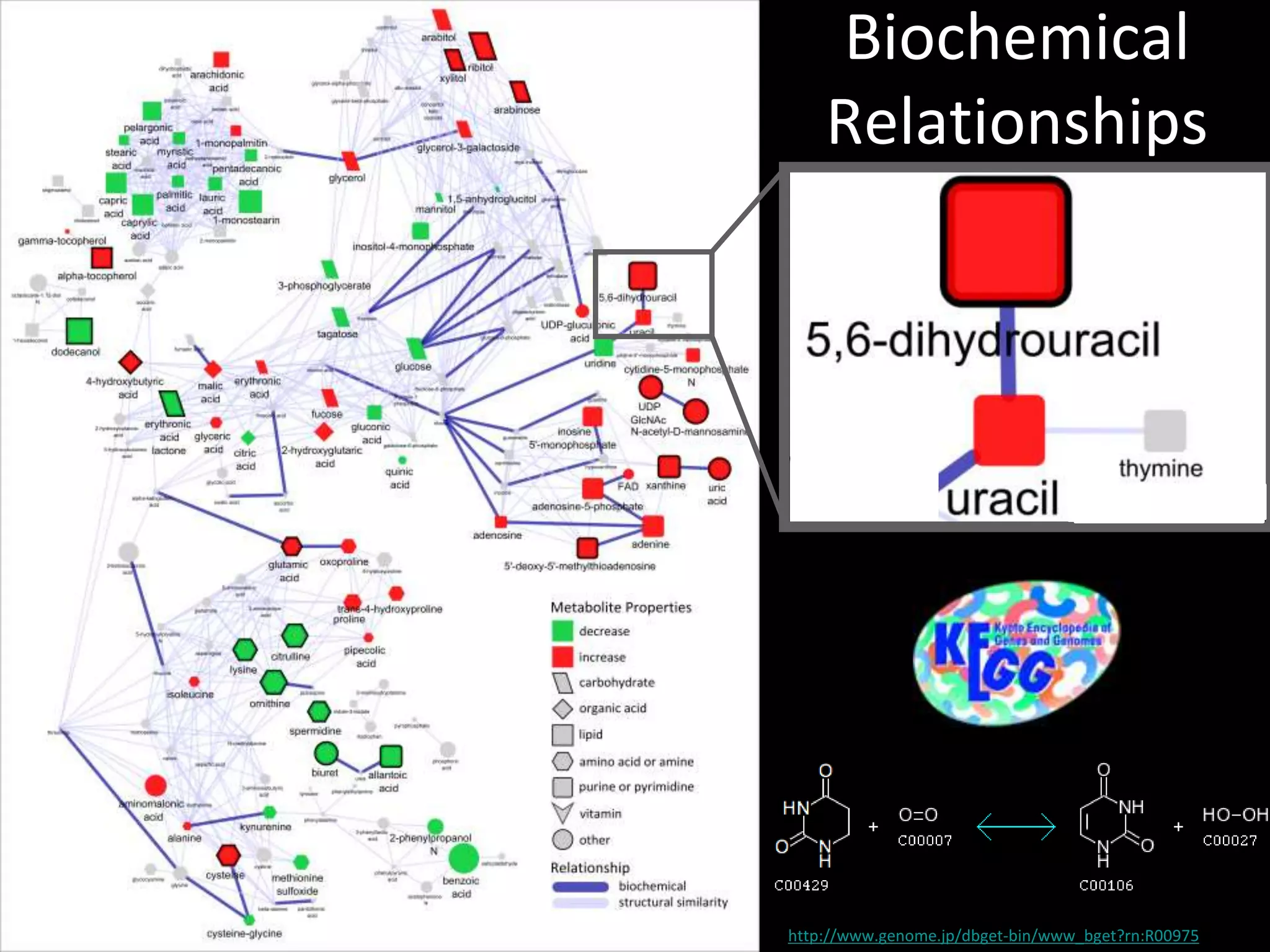
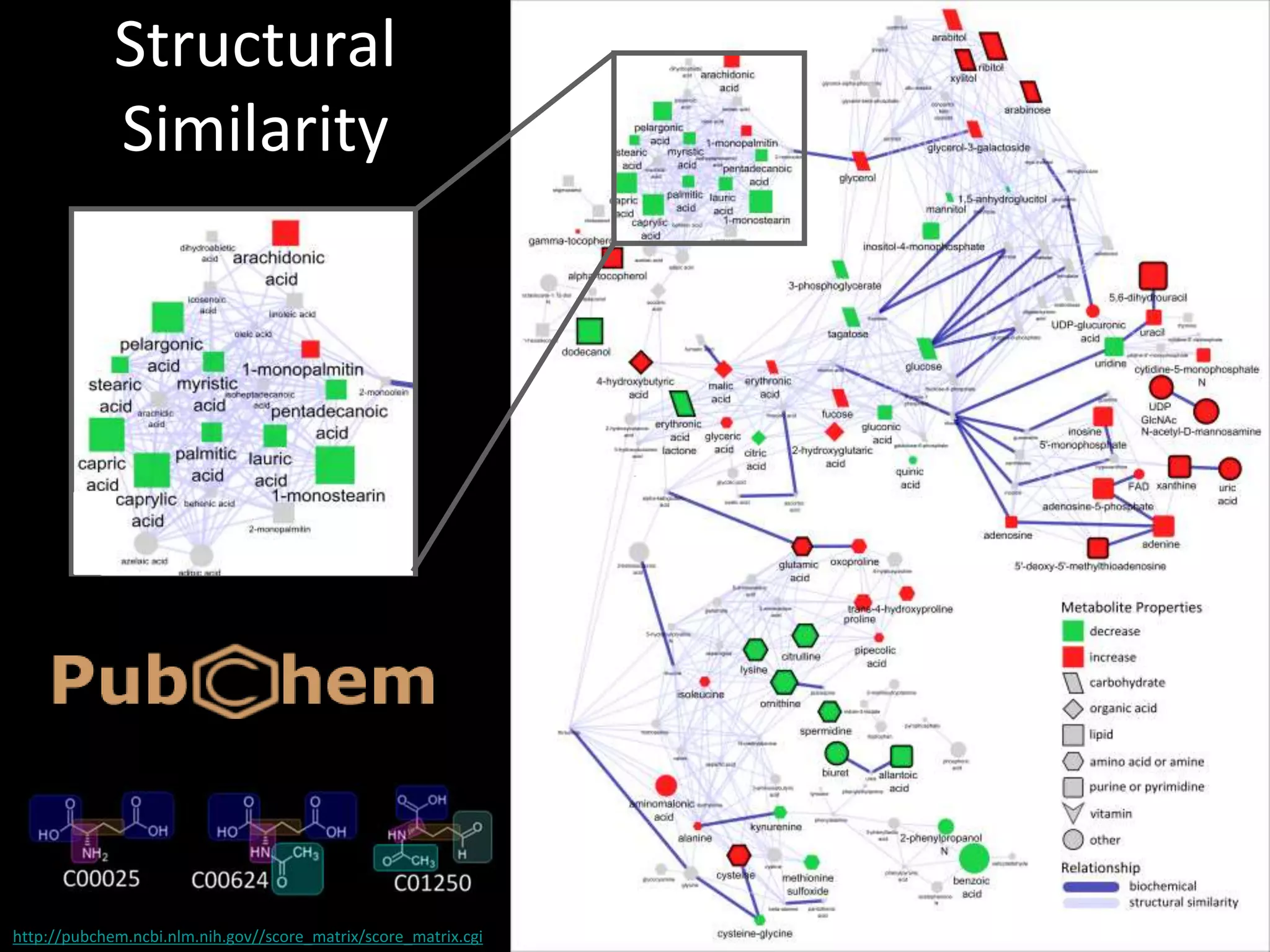
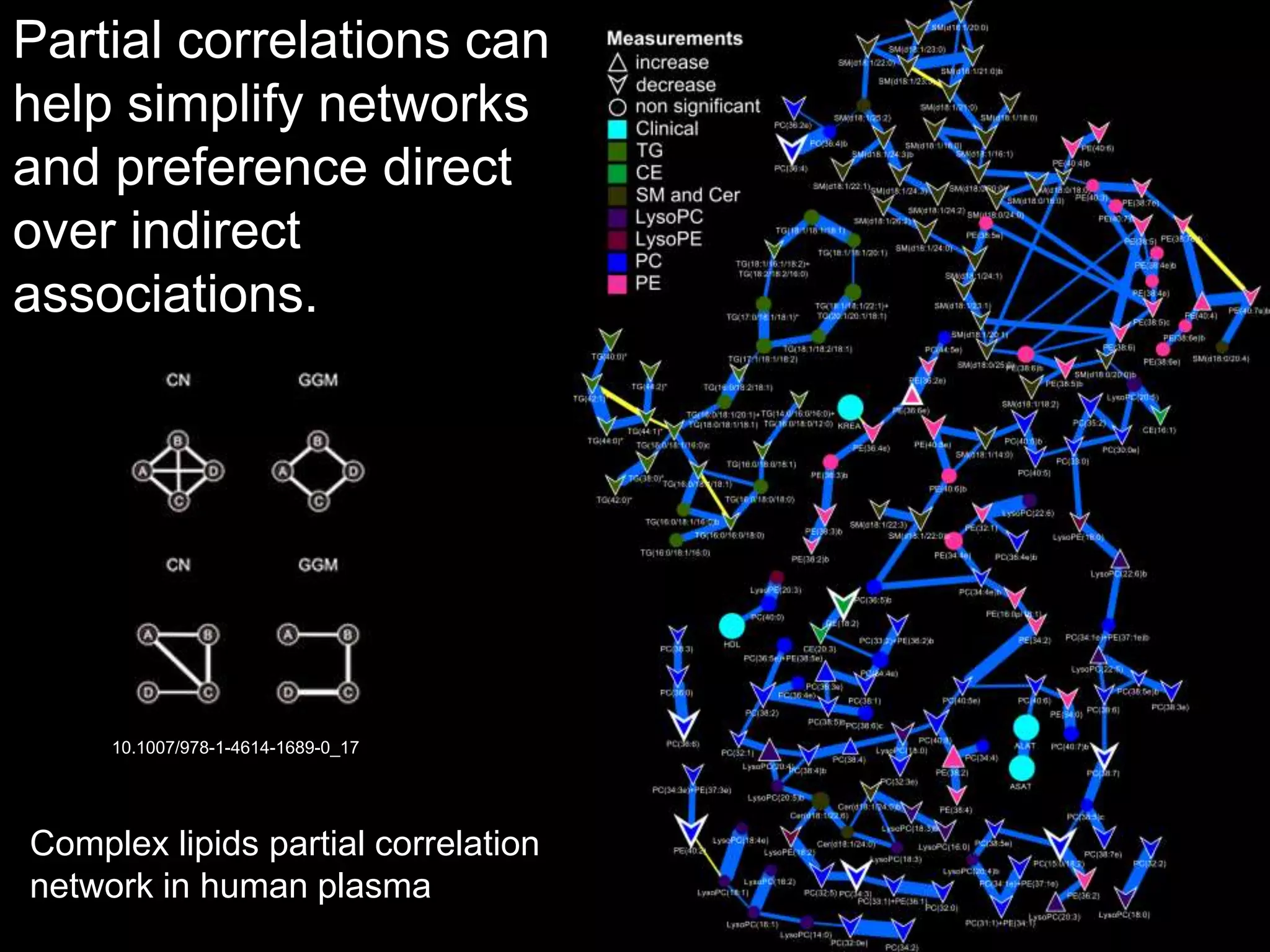
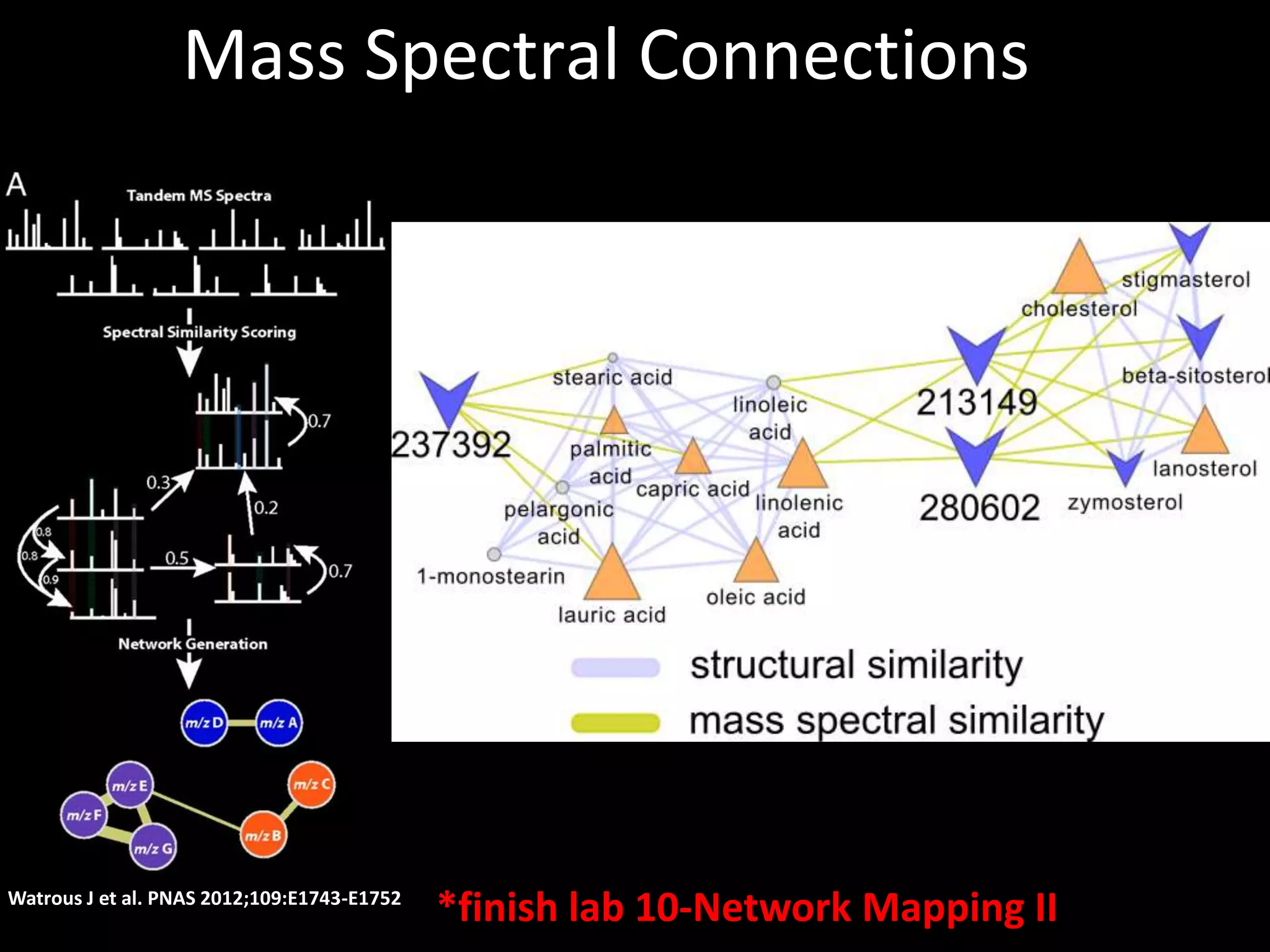
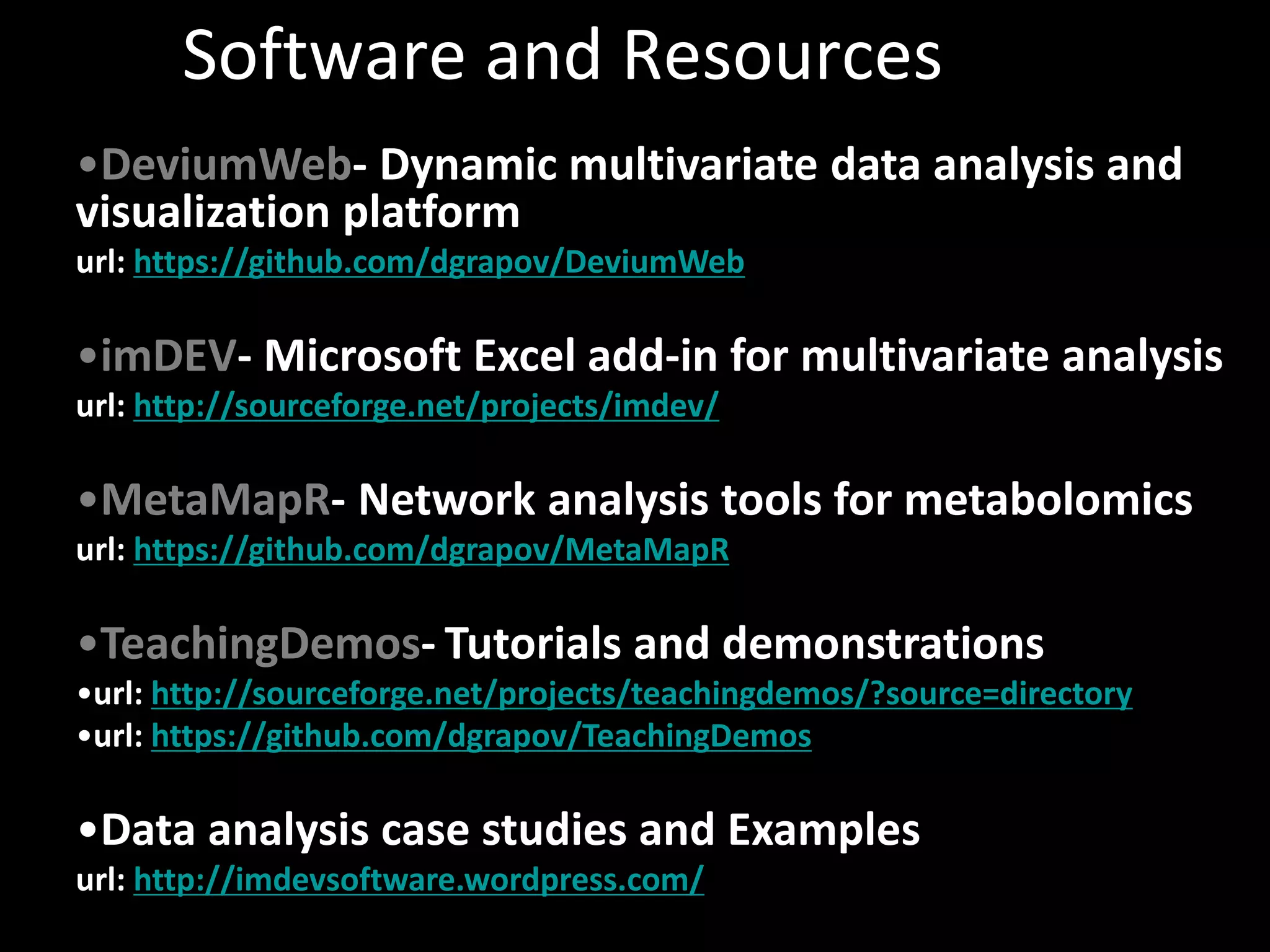
This document provides an introduction and overview of tutorials for metabolomic data analysis. It discusses downloading required files and software. The goals of the analysis include using statistical and multivariate analyses to identify differences between sample groups and impacted biochemical domains. It also discusses various data analysis techniques including data quality assessment, univariate and multivariate statistical analyses, clustering, principal component analysis, partial least squares modeling, functional enrichment analysis, and network mapping.