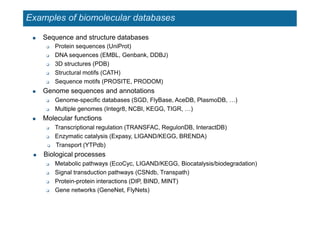
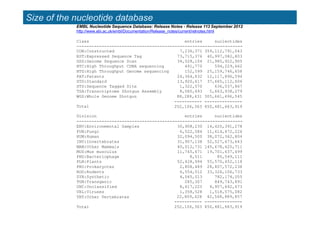
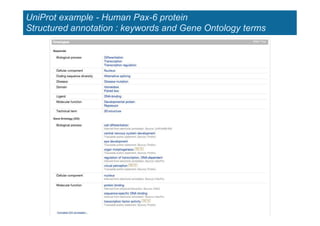
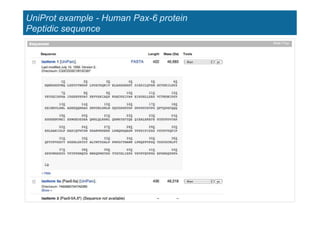
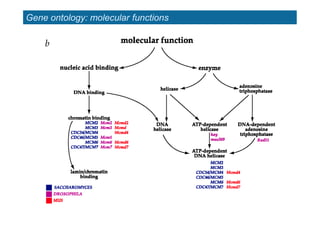
The document provides an overview of several important biomolecular databases. It begins by describing examples of biological databases including nucleic acid sequence databases like GenBank, EMBL, and DDBJ that contain DNA and RNA sequences. It also mentions UniProt, the major database of protein sequences and annotations. Finally, it briefly introduces the Gene Ontology project, which provides controlled vocabularies to describe gene and gene product attributes.