JBEI Research Highlights September 2016
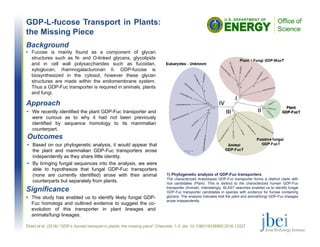
- 1. GDP-L-fucose Transport in Plants: the Missing Piece Outcomes • Based on our phylogenetic analysis, it would appear that the plant and mammalian GDP-Fuc transporters arose independently as they share little identity. • By bringing fungal sequences into the analysis, we were able to hypothesize that fungal GDP-Fuc transporters (none are currently identified) arose with their animal counterparts but separately from plants. Ebert et al. (2016) “GDP-L-fucose transport in plants: the missing piece” Channels. 1-3. doi: 10.1080/19336950.2016.12227 Background • Fucose is mainly found as a component of glycan structures such as N- and O-linked glycans, glycolipids and in cell wall polysaccharides such as fucoidan, xyloglucan, rhamnogalacturonan II. GDP-fucose is biosynthesized in the cytosol, however these glycan structures are made within the endomembrane system. Thus a GDP-Fuc transporter is required in animals, plants and fungi. Significance • This study has enabled us to identify likely fungal GDP- Fuc homologs and outlined evidence to suggest the co- evolution of this transporter in plant lineages and animals/fungi lineages. 1) Phylogenetic analysis of GDP-Fuc transporters The characterized Arabidopsis GDP-Fuc transporter forms a distinct clade with rice candidates (Plant). This is distinct to the characterized human GDP-Fuc transporter (Animal). Interestingly, BLAST searches enabled us to identify fungal GDP-Fuc transporter candidates in species with evidence for fucose containing glycans. The analysis indicates that the plant and animal/fungi GDP-Fuc lineages arose independently. Approach • We recently identified the plant GDP-Fuc transporter and were curious as to why it had not been previously identified by sequence homology to its mammalian counterpart.
- 2. 13C Metabolic Flux Analysis for systematic overproduction of fatty acids in yeast Outcomes • Expression of ATP citrate lyase (ACL) to increase acetyl-CoA does not increase fatty acid production significantly (Fig. 1). • Flux profiles from 2S 13C MFA show that the extra acetyl-CoA produced by ACL goes into the malate synthase (MALS) rather than fatty acid production (ACCOACr, Fig. 2). • Downregulating MALS and knocking out glycerol production (GPD1), increased fatty acid production by ~70% (Fig. 1). 1) Fatty acid production for the various strains studied in this manuscript The addition of ACL was expected to increase acetyl-CoA availability but did not increase final production. However, the downregulation of MLS did increase production, as suggested by flux analysis. The highest production was obtained by knocking out glycerol production, improving production in the overall engineering process by 70%. Ghosh et al. (2016) “13C Metabolic Flux Analysis for Systematic Metabolic Engineering of S. cerevisiae for Overproduction of Fatty Acids” Front. Bioeng. Biotechnol. 4:76. doi: 10.3389/fbioe.2016.00076 Background • Tools for systematic and efficient redirection of microbial metabolism into the abundant production of desired bioproducts are a need in metabolic engineering. • Previous engineering attempts with S. cerevisaie led to a production of free fatty acids at a titer of 400 mg/L. Significance • The use of 2S 13C MFA provides a way to ascertain the fate of carbon from feed to product and produces new insights to improved biofuel yield in a systematic fashion. 2) Cytosolic acetyl-CoA balances obtained from 2S-13C MFA Flux of acetyl-CoA (accoa) producing and consuming reactions is shown as a sankey diagram. Reactions on the left of the diagram produce acetyl-coA and reactions on the right consume it, with arrow sizes indicating total flux. Once the ACL is added (WRY2 ACL), total acetyl-coA flux increases, but this extra flux flows into MALS, which is consistent with the lack of fatty acid production increase (Fig. 1). Approach • We combined 13C labeling data with comprehensive genome-scale models (2S 13C MFA) to shed light onto microbial metabolism and improve metabolic engineering efforts.
- 3. Proteomic Characterization of Golgi Membranes Enriched from Arabidopsis Suspension Cell Cultures Outcomes • A detailed protocol for isolating Golgi membranes with various ‘tricks’ developed at JBEI has been outlined • The method yields about 26 % Golgi proteins and in combination with informatics approaches the method could be used for proteomic surveys in plants Hansen et al. (2016) "Proteomic Characterization of Golgi Membranes Enriched from Arabidopsis Suspension Cell Cultures". Methods Mol Biol, 1496, 91-109. doi, 10.1007/978-1-4939-6463-5_8 Background • The stacked membrane structures of the Golgi apparatus along with its interactions with the cytoskeleton and ER have historically made the isolation and purification of this organelle difficult. Density centrifugation has typically been used to enrich Golgi membranes from plant microsomal preparations, and aside from minor adaptations, the approach is still widely employed. Significance • Significant effort has been expended at JBEI to develop these methods and this published protocol attempts to distil the ‘tricks’ for the plant science community. 1) Schematic outlining the enrichment of Golgi membranes by density centrifugation Approach • We outline a detailed protocol that we have extensively refined for the enrichment of Golgi membranes from Arabidopsis suspension cell cultures. The approach is suitable for samples that can be used for downstream processes such as comparative proteomic studies. 2) Validation of Golgi proteins can be undertaken by localization with fluorescently tagged proteins This validation process was also outlined in the published protocol. The Golgi marker aManI-mCherry is shown as a Golgi reference signal. The UXT3-YFP proteins clearly localizes to the Golgi (see merge).
- 4. Cell Wall Composition and Candidate Biosynthesis Gene Expression During Rice Development Outcomes • Through analysis of the correlations within and among the cell wall and gene expression datasets, we establish testable hypotheses for the potential interactions among cell wall polymers, and the functions of putative cell wall synthesis genes • This work successfully moves beyond the observation that particular GTs and ATs are grass-diverged and highly expressed to providing testable hypotheses for specific cell wall synthesis activities, pinpointing nine uncharacterized genes with putative functions in rice cell wall synthesis. Lin et al. (2016) "Cell Wall Composition and Candidate Biosynthesis Gene Expression During Rice Development". Plant Cell Physiol. doi, 10.1093/pcp/pcw125 Background • Rice straw comprises almost one quarter of all agricultural waste globally • The development of rice is continuous and there is not a distinct juvenile to adult boundary • Significant knowledge gaps exist about the roels of cell wall biosynthesis genes in rice correlated to changes in composition. Significance • These results may lead to the discovery of novel grass cell wall synthesis enzymes, and may improve biofuel production and other agricultural uses of cereals and grasses. Approach • We have developed a cell wall composition atlas that provides a baseline for evaluating the variation of cell wall composition across developmental time points and organs in rice, and for comparisons to other plant species. Abundance of cell wall components and properties measured by chemical assays. Color intensity indicates the z-score of abundance for each component. Red indicates relative enrichment and blue, relative scarcity. Slashed squares are missing values due to insufficient sample mass. The mean, un-normalized mass for each component is shown above the heatmap. The color ribbons on the left of heatmap indicate organs.
- 5. Towards Economically Sustainable Lignocellulosic Biorefineries Outcomes • Given different production pathways for MA and the market price volatility of MA, multiple scenarios were constructed (Fig. 2) • Overall, higher MA price scenarios (‘red’) are more favorable. • Minimizing the energy costs associated with the aerobic fermentation can make the process more attractive from an economical standpoint Konda et al. (2016) “Towards economically sustainable lignocellulosic biorefineries”, In Valorization of Lignocellulosic Biomass in a Biorefinery: From Logistics to Environmental and Performance Impact. Nova publishers. Background • Economic viability is essential for the commercialization of cellulosic biorefineries for the production of advanced biofuels • Due to the low market price of fuels and market price volatility, cellulosic biofuel production faces challenging economics • Co-production of value added chemicals, alongside of biofuel, can potentially enable cost-competitive production of biofuels • Among various co-production strategies (Fig. 1), feedstock engineering to incorporate easily extractible chemicals/intermediates is a potential option Significance • This study has quantified the potential impact of the co-production of chemicals from a technoeconomic standpoint. Results help researchers to design and develop processes that meet commercially relevant targets for key performance metrics (e.g., yield) Approach • To investigate the impact of co-products on the production cost of biofuel, we have considered the case of engineered sweet sorghum for the co- production of Muconic Acid (MA), a precursor for Adipic Acid. • To facilitate economic analysis, detailed biorefinery model was developed in a commercial software platform SuperPro Designer 2) Estimated min. ethanol selling price (MESP). The ‘blue’ and ‘red’ bars represent different market scenarios with regards to MA selling price (i.e., $1000/MT and $1500/MT, respectively). S1 (no co-product), S2 (sugars- to-MA pathway), S3 (PCA-to-MA pathway), S4 (MA from both sugars and PCA) 1) A simplified superstructure representation of integrated cellulosic biorefineries for the co-production of chemicals together with biofuel.
- 6. Development of Next Generation Synthetic Biology Tools for S. venezuelae Outcomes • We generated 45 expression vectors that can simultaneously integrate at three different attB sites and characterized 15 promoters that possess relative strengths that vary over 3 orders of magnitude. • The tools and information generated were used to develop a chromosomally integrated bisabolene production pathway that produced 5.4 mg/L. Phelan et al. (2016) “Development of Next Generation Synthetic Biology Tools for use in Streptomyces venezuelae” ACS Synth Biol., doi: 10.1021/acssynbio.6b00202 Background • Streptomyces have a rich history as producers of important natural products, and this genus of bacteria has recently garnered attention for its potential applications in the broader context of synthetic biology. • The dearth of genetic tools available to control and monitor protein production precludes rapid and predictable metabolic engineering Significance • These results help establish a toolbox that will streamline the introduction or modification of biosynthetic pathways in S. venezuelae by allowing more predictable protein production and simplifying the introduction of multiple genes or operons relevant to biofuel production. Approach • We developed a suite of standardized, orthogonal integration vectors and an improved method to monitor protein production in this host • These tools were then applied to characterize heterologous promoters and various attB chromosomal integration sites, coupled with a demonstration of producing the biofuel precursor bisabolene using a chromosomally integrated expression system. Small scale screening of bisabolene producing strains Screen of seven differing fluorescent proteins integrated at the VWB attB site under the control of strong, constitutive glyceraldehyde-3-phosphate dehydrogenase promoter (Pgapdhp(EL)) from Eggerthella lenta
- 7. Approaches to Understanding Protein Hypersecretion in Fungi Outcomes • Case studies: (1) amylase production in Aspergillus niger, (2) cellulase secretion in Trichoderma reesei, and (3) secretion in Neurospara crassa. • Traditional methods of isolation and improvement of protein production hosts, such as mutagenesis and screening, and reverse genetics to demonstrate the action of specific genes, will continue to have value for the future generation of improved strains. • The secretion pathway itself is key, as UPR and the cell’s secretion machinery play important roles in the level of protein output from the cell. • The impact of the advances of the genomics era is that these standard approaches have become more powerful and the rate of progress has increased dramatically Reilly et al. (2016) “Approaches to understanding protein hypersecretion in fungi” Fungal Biology Reviews, doi: 10.1016/j.fbr.2016.06.002 Background • Fungi play an incredibly important role in industrial biotechnology, and applications that utilize fungal enzymes have grown to a much larger number of industrial applications, from food and beverage production to the digestion of plant biomass in bioenergy and bioproducts applications • In this review paper, we discuss three “case studies” in the area of fungal protein hyperproduction Significance • The combination of traditional and advanced approaches will enable the development of new and better fungal protein production hosts for missions in bioenergy and beyond. Aspergillus niger Neurospara crassa
- 8. Dynamic Changes of Substrate Reactivity and Enzyme Adsorption on Partially Hydrolyzed Cellulose Outcomes • Results suggest that cellobiohydrolase CBH1 (Cel7A) and endoglucanases EG2 (Cel5A) adsorption capacities decreased as cellulose was progressively hydrolyzed, likely due to the “depletion” of binding sites; while the degree of synergism between CBH1 and EG2 varied depending on the enzyme loading and the substrates. • Moreover, these results suggest that more oligosaccharides (DP>3 cellodextrins) than soluble sugars could be produced during cellulose hydrolysis and influenced by both cellulose substrate and enzyme loadings and combinations. Shi et al. (2016) "Dynamic changes of substrate reactivity and enzyme adsorption on partially hydrolyzed cellulose" Biotechnol Bioeng, doi, 10.1002/bit.26180 Background • The enzymatic hydrolysis of cellulose is a thermodynamically challenging catalytic process that is influenced by both substrate-related and enzyme- related factors. The limited understanding of hydrolysis reaction mechanisms and factors controlling hydrolysis effectiveness impede many promising applications in the real world. Approach • A proteolysis approach was applied to recover and clean the partially converted cellulose at the different stages of enzymatic hydrolysis to monitor the hydrolysis rate as a function of substrate reactivity/accessibility and investigate surface characteristics of the partially converted cellulose. Significance • This study provides a better understanding of the relationship between dynamic change of substrate features and the functionality of various cellulases during enzymatic hydrolysis.
- 9. A Robust Gene Stacking Method utilizing Yeast Assembly for Plant Synthetic Biology Outcomes • We have demonstrated how our method can be applied to a number of plant metabolic engineering efforts. • We have built a publicly available library of over 100 plant DNA parts to aid other plant synthetic biology efforts. Background • Plant synthetic biology is a young field that has the potential to address many sustainability, bioenergy, and agricultural issues • There has been a dearth of DNA parts libraries, versatile transformation vectors, and efficient assembly strategies for plant transformations. Approach • We have developed a method that utilizes yeast homologous recombination to assemble DNA into plant transformation vectors. Significance • Our approach provides an alternative method to efficiently assembling DNA into plant transformation vectors. • Improved DNA assembly will enable more complex plant metabolic engineering efforts to manipulate plant feedstocks to be more sustainable and have better bioenergy traits. 2) Yeast homologous recombination vectors enable the stacking of multiple genes together. Gene stacking can enable yield from metabolic engineering efforts or the introduction of entirely heterologous pathways. 1) Hierarchical assembly of plant DNA parts for plant synthetic biology. A library of DNA parts (Level 0) can be efficiently assemble together into gene cassettes. These cassettes can be stitched together via yeast homologous recombination. Shih et al. (2016) “A robust gene stacking method uti.izing yeast assembly for plant synthetic biology” Nature Communications. doi: 10.1038/ncomms13215
- 10. Glycosylation of Inositol Phosphorylceramide Sphingolipids is Required for Normal Growth and Reproduction in Arabidopsis Outcomes • Assignment of a direct role for GIPC glycan headgroups in the impaired processes in iput1 mutants is complicated by the vast compensatory changes in the sphingolipidome. • However, our results reveal that the glycosylation steps of GIPC biosynthesis are important regulated components of sphingolipid metabolism. 1) Rescued iput1 mutants are dwarfed and have oxidative stress. Plants from two different transformant lines are shown in comparison to wild type. The leaves of the rescued plants show lesions indicating oxidative stress (as confirmed with DAB staining, not shown). The plants also have high levels of salicylic acid. Tartaglio et al. (2016) "Glycosylation of inositol phosphorylceramide sphingolipids is required for normal growth and reproduction in Arabidopsis". Plant J. doi, 10.1111/tpj.13382 https://www.ncbi.nlm.nih.gov/pubmed/27643972. Background • Sphingolipids are a major component of plant plasma membranes and endomembranes, and mediate a diverse range of biological processes, including cellulose biosynthesis and plant-microbe interactions. • Study of the highly glycosylated glycosyl inositol phosphorylceramide (GIPC) sphingolipids, has been slow due to challenges associated with GIPCs' extractability, and their functions in the plant remain poorly characterized. Significance • This study corroborates previously suggested roles for GIPC glycans in plant growth and defense, suggests important roles for them in reproduction, and demonstrates that the entire sphingolipidome is sensitive to their status. 3) Sphigolipidomics of rescued iput1 mutants. The mutants show accumulation of IPC, the substrate for IPUT1 glucuronosyltransferase. The fully glycosylated GIPC is strongly reduced in the mutants. Other changes in ceramides are pleiotropic effects of the primary defect in GIPC biosynthesis. Approach • We recently discovered an Arabidopsis GIPC glucuronosyltransferase, INOSITOL PHOSPHORYL-CERAMIDE GLUCURONOSYLTRANSFERASE 1 (IPUT1), which is the first enzyme in the GIPC glycosylation pathway. • Using a pollen-specific rescue construct, we have here isolated homozygous iput1 mutants. 2) Pollen from iput1 plants has very low transmission efficiency. Pollen tube growth appears normal, but the mutant pollen tubes cannot efficiently be guided to the micropyle. The few efficient pollinations do not lead to viable embryos.
- 11. XA-21-specific Induction of Stress-related Genes Following Xanthomonas Infection of Detached Rice Leaves Outcomes • We identified eight genes that are up-regulated in both in elf18 treated EFR:XA21:GFP rice leaves and Xoo infected XA21 rice leaves. • Our results show that even though the EFR:XA21:GFP-mediated response does not confer robust resistance to Xoo, similar genes are up-regulated during both EFR:XA21:GFP- and Myc:XA21- mediated response Thomas et al. (2016) “XA-21-specific Induction of Stress-related Genes Following Xanthomonas Infection of Detached Rice Leaves” PeerJ, 4, e2446 doi: 10.7717/peerj.2446 Background • Plant immunity is mediated, in part, by cell surface immune receptors that recognize molecules produced by microbes.. • Studies of the XA21-mediated immune response have been limited by the lack of rapid assays and well-characterized genetic markers. Significance • The discovery of RaxX and the establishment of the detached leaf infection assay described here provide useful tools for studying XA21-mediated immunity that will improve bioenergy crop productivity. Approach • We developed a detached leaf infection assay to quickly and reliably measure activation of the XA21-mediated immune response using genetic markers. • The detached leaf infection assay allows a more uniform distribution, compared to the scissor inoculation method • We used RNA sequencing of elf18 treated EFR:XA21:GFP plants to identify candidate genes that could serve as markers for XA21 activation. The stress-related marker gene PR10b is up-regulated in Xanthomonas infected XA21 rice. PR10b expression in detached Kitaake and Myc:XA21 rice leaves with 10 mM MgCl2 mock treatment or infected with PXO99A or PXO99A1hrpA1 (1hrpA1) at an O.D.600 of 0.1. Letters represent statistically significant differences between mean expression values (p < 0:05) determined by using a TukeyKramer HSD test. This experiment was repeated three times with similar results.
- 12. Comprehensive in vitro Analysis of Acyltransferase Domain Exchanges in Modular Polyketide Synthases and its Application for Short-chain Ketone Production Significance • These results greatly enhance the mechanistic understanding of modular PKS systems. • As demonstrated in short-chain ketone production, our results prove the benefits of using engineered PKSs as a synthetic biology tool for chemical production. Yuzawa et al. (2016) “Comprehensive in vitro analysis of acyltransferase domain exchanges in modular polyketide synthases and its application for short-chain ketone production” ACS Synth Biol doi: 10.1021/acssynbio.6b00176. Background • Modular polyketide synthases (PKSs) are polymerases that utilize acyl-CoA as substrates. • Each polyketide elongation reaction is catalyzed by a set of protein domains called a module. • Individual acyltransferase (AT) domains in the PKS modules determine the specific acyl-CoA incorporated into each polyketide condensation reaction. • Although successful exchange of individual AT domains lead to the biosynthesis of a large variety of novel compounds, hybrid PKSs often show significantly decreased activities. Approach • Using model PKS systems, we have systematically analyzed the segments of AT domains and associated linkers in AT exchanges in vitro. Outcomes • We have identified the boundaries within a module that can be used to exchange AT domains while maintaining protein stability and enzyme activity. • The optimized domain boundary is highly conserved, which facilitates AT domain replacement in most PKS modules. New synthetic biology tool! etc… Acceptor PKS AT-swapped mutant library KAL PAL1 PAL2 Reduction loop ACP Reduction loop ACP KAL PAL1 PAL2 KS AT KS AT Donor PKS KAL PAL1 PAL2 Reduction loop ACPKS AT
- 13. Outcomes • A comprehensive phylogenetic tree of Rack1 proteins has been constructed. • The current research status on the Rack1 protein has been summarized. • Future research areas regarding Rack1 are indicted. Zhang et al. (2016) “Roles of Rack1 proteins in fungal pathogenesis” BioMed Res Int. http://dx.doi.org/10.1155/2016/4130376 Background • Fungal pathogens cause severe losses of feedstock biomass. • The conserved Rack1 protein from plant pathogenic fungi is a potential target for developing effective fungicides. Significance • This study reveals the conserved and dynamic role of Rack1 proteins in different fungal species. • This study facilitates the development of fungicides targeting the fungal Rack1 protein to improve biofuel feedstock production. Approach • Phylogenetic analysis • Compare functions of Rakc1 protein from different organisms Roles of Rack1 Proteins in Fungal Pathogenesis
- 14. From Lignin Association to Nano-/Micro-particle Preparation: Extracting Higher Value of Lignin Outcomes • Literature studies on preparation, functionalization and application of lignin nanoparticles are still limited. • Fundamental understandings of lignin solution structures, their relation to nanoparticle preparation and properties, nanoparticles stability and dispersion will certainly lead to advancement in synthesizing lignin-based functional materials. • From an industrial point of view, it is desirable to minimize utilization of hazardous solvents or to replace them with green solvents such as water, supercritical CO2 and room temperature ionic liquids (RTILs), etc. Zhao et al. (2016) “From Lignin Association to Nano-/Micro-particle Preparation: Extracting Higher Value of Lignin” Green Chemistry, doi: 10.1039/c6gc01813k Background • Value-added applications of lignin are essential for economic viability for future bio-refineries. • Conversion of lignin stream to value-added components still remains a challenge which casts a shadow over the fate of future biorefineries Significance • This work provides a perspective that would prompt the development of new processes for manufacturing lignin nanoparticles with designed properties. Approach • Lignin nanoparticles emerge as novel building blocks with unique and tunable properties, which is expected to play a vital role in value-added applications. • Methods for preparation of lignin nanoparticles are essentially adopted from those used in manufacturing polymer nanoparticles.