JBEI Research Highlights - January 2018
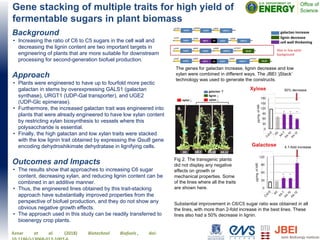
- 1. Gene stacking of multiple traits for high yield of fermentable sugars in plant biomass Background • Increasing the ratio of C6 to C5 sugars in the cell wall and decreasing the lignin content are two important targets in engineering of plants that are more suitable for downstream processing for second-generation biofuel production. Approach • Plants were engineered to have up to fourfold more pectic galactan in stems by overexpressing GALS1 (galactan synthase), URGT1 (UDP-Gal transporter), and UGE2 (UDP-Glc epimerase). • Furthermore, the increased galactan trait was engineered into plants that were already engineered to have low xylan content by restricting xylan biosynthesis to vessels where this polysaccharide is essential. • Finally, the high galactan and low xylan traits were stacked with the low lignin trait obtained by expressing the QsuB gene encoding dehydroshikimate dehydratase in lignifying cells. Outcomes and Impacts • The results show that approaches to increasing C6 sugar content, decreasing xylan, and reducing lignin content can be combined in an additive manner. • Thus, the engineered lines obtained by this trait-stacking approach have substantially improved properties from the perspective of biofuel production, and they do not show any obvious negative growth effects. • The approach used in this study can be readily transferred to bioenergy crop plants. Aznar et al. (2018) Biotechnol Biofuels , doi: The genes for galactan increase, lignin decrease and low xylan were combined in different ways. The JBEI ‘jStack’ technology was used to generate the constructs. Fig 2. The transgenic plants did not display any negative effects on growth or mechanical properties. Some of the lines where all the traits are shown here. Substantial improvement in C6/C5 sugar ratio was obtained in all the lines, with more than 2-fold increase in the best lines. These lines also had a 50% decrease in lignin.
- 2. Constraining genome-scale models to represent the bow tie structure of metabolism Background • Mapping metabolic flux is critical for efficient engineering of microbes for the production of biofuels and bioproducts. • Our approach is based on simplifying approximation that metabolic flux from peripheral metabolism into central “core” carbon metabolism is minimal, and can be omitted when modeling isotopic labeling in core metabolism. • Validity of this “two-scale” or “bow tie” approximation is supported both by the ability to accurately model experimental isotopic labeling data, and by experimentally verified metabolic engineering predictions using these methods. Approach • Systematically calculate flux bounds for any specified “core” of a genome-scale model so as to satisfy the bow tie approximation • Automatically identify an updated set of core reactions that can satisfy this approximation more efficiently. • Ran both algorithms on the iJR904 E. coli genome scale model, using exchange and biomass fluxes from previously published sample data. Outcomes and Impacts • These methods accelerate and automate the identification of a biologically reasonable set of core reactions for use with 13 C MFA or 2S-13 C MFA. • These methods accelerate and automate the identification of a biologically reasonable set of core reactions. • We have dan open source Python implementation of these algorithms at https://github.com/JBEI/limitfluxtocore that is available to the broader community. Backman et al. (2018) Metabolites, doi: 10.3390/metabo8010003 The bow tie structure of metabolism. The bow tie structure of metabolism describes how numerous different types of carbon sources (left side of figure) are funneled into a small set of precursor metabolites (middle of figure) in central carbon metabolism that are subsequently processed into a large set of peripheral metabolites (right side of figure).
- 3. Employing a biochemical protecting group for a sustainable indigo dyeing strategy Background • 50,000 tons of indigo are synthesized annually, of which 95% is used to dye the over 4 billion denim garments manufactured annually. • Indigo is chemically synthesized on an industrial scale from aniline, a toxic compound derived from the petroleum product benzene. • There is a compelling need for an environmentally sustainable approach that achieves production of indigo as a valuable bioproduct. Approach • Developed a sustainable dyeing strategy that not only circumvents the use of toxic reagents for indigo chemical synthesis but also removes the need for a reducing agent for dye solubilization. • This alternative indigo dyeing process that mimics the natural biochemical protecting group strategy employed by the Japanese indigo plant P. tinctorium. • Goal was to gain temporal control over the regeneration of the indoxyl intermediate to achieve indigo crystallization in the cotton fibers. Outcomes and Impacts • We identified the gene sequence of a novel glucosyltransferase from P. tinctorium (PtUGT1) with activity on indoxyl, characterized the enzyme structurally, and successfully co-expressed it with the oxygenase FMO in E. coli to produce up to 2.9 g/L indican in vivo from fed tryptophan. • This bioindican was shown to be an effective fabric dye when hydrolyzed by a β-glucosidase. Hsu et al. (2018) Nature Chemical Biology , doi: 10.1038/nchembio.2552 Overall GT-B fold of PtUGT1 (PDB ID 5NLM). The N-terminal Rossmann domain (blue) consists of a seven-stranded parallel β-sheet surrounded by nine α-helical segments, and the C-terminal Rossmann domain (red) consists of a six-stranded parallel β-sheet and five α-helices. (a) Top row, Pure indican with no β-glucosidase (BGL); pure indican with β-glucosidase; bio-indican with β-glucosidase. Bottom row, Indigo, non-reduced; indigo, reduced with sodium dithionite. (b) Scarf (100% white cotton gauze fabric) dyed with indican.
- 4. Forward genetics screen coupled with whole-genome resequencing identifies novel gene targets for improving heterologous enzyme production in Aspergillus niger Background • Efficient expression of recombinant enzymes capable of hydrolyzing polysaccharides present after pretreatment is a critical need at JBEI. • Aspergillus niger is a promising recombinant host under development at JBEI to meet this need. • We have previously identified several enzymes capable of maintaining high activity in the presence of ionic liquids used in biomass pretreatment that we would like to express in A. niger. Approach • We developed a forward genetics screen coupled with whole-genome resequencing to identify specific lesions responsible for a protein hyper-production phenotype in A. niger. • Use bioinformatics to identify loci associated with heterologous enzyme hyper-production, including those that appear to be specific to the heterologous protein expression construct used. Outcomes and Impacts • Our strategy successfully identified novel targets, including a low-affinity glucose transporter, MstC, whose deletion significantly improved secretion of recombinant proteins driven by a glucoamylase promoter. Deletion of mstC loci improved heterologous enzyme production driven by a glucoamylase promoter (PglaA) fourfold. • Mutations in the Ypt/Rab GTPase-activating protein found in BOTH strain J03 2.8 and NAT strain J03 4.3 might further increase production of heterologous BG if introduced into one HET strain. Reilly et al. (2018) Applied Microbiology and Biotechnology , 102(4), 1797-1807, doi: 10.1007/s00253-017-8717-3 Mutations in J03-derivative strains associated with hyper-production of heterologous and/or native beta-glucosidase. Loss of the mstC locus enhances heterologous enzyme production.
- 5. Isolation and characterization of novel mutations in the pSC101 origin that increase copy number Background • Synthetic biology is predicated on predictable levels of gene expression. This is often achieved through the use of plasmids with specific copy numbers. • Isolating novel copy number variants, and precisely characterizing their expression profiles may enable finer tuning of synthetic genetic circuits. Approach • To Identify high copy variants of the pSC101 origin, we leveraged random mutagenesis and plate based selections. • We modelled the location of the most common mutations to predict other potential residues that control copy number. • To evaluate the relationship between copy number and fluorescent protein production strength we created site-directed mutants within pSC101 vectors expressing RFP controlled by an arabinose-inducible promoter. Outcomes and Impacts • We identified 10 previously unidentified mutations within the pSC101 origin that increase plasmid copy. Theses residues are likely to contribute to RepA dimerization. • We identified two specific mutations within the RepA protein that result in copy numbers comparable to other commonly used medium and high copy vectors in Escherichia coli. • By combining mutations we were able to create super-high copy vectors, with copy numbers reaching ~500 copies/cell. Thompson et al. (2018) Sci Rep, doi: 10.1038/s41598-018-20016-w
- 6. Engineering E. coli for simultaneous glucose-xylose utilization during methyl ketone production Outcomes and Impacts • Two strategies were effective at enhancing methyl ketone titer: (1) chromosomal deletion of pgi to increase intracellular NADPH supply and (2) downregulation of CRP (cAMP receptor protein) by replacement of the native RBS. • Combining these strategies resulted in the most favorable overall phenotypes (see strains XW1074 and especially XW1075 in figure at right). • This work demonstrated a strategy for engineering simultaneous utilization of C6 and C5 sugars in E. coli without sacrificing production of methyl ketones (diesel fuel blending agents) from cellulosic biomass. Wang et al. (2018) Microbial Cell Factories , doi: 10.1186/s12934-018-0862-6 Background • We previously developed an E. coli strain that efficiently converts glucose to methyl ketones (MK) for use as diesel fuel blending agents. • Since lignocellulosic biomass contains a substantial portion of xylose (typically the second most abundant sugar after glucose), we re-engineered this strain to more efficiently metabolize sugars by facilitating simultaneous, rather than the native sequential (diauxic), utilization of glucose and xylose. Approach • Genetic manipulations were performed to alleviate carbon catabolite repression in our most efficient MK-producing strain (EGS1895). • A strain engineered for constitutive expression of genes involved in xylose transport and metabolism showed synchronized C6-C5 sugar consumption, but a 5-fold decrease in MK production. Further efforts were made to improve MK production in this strain (XW1014). . Summary comparison of methyl ketone production and sugar consumption for engineered strains. Methyl ketone yield, methyl ketone productivity, and sugar consumption period are each normalized to the maximum value among the six strains. Strain XW1075 performed best overall.
- 7. Biomass pretreatment using deep eutectic solvents (DESs) from lignin derived phenols Outcomes and Impacts • Demonstrated that novel DESs formed from a renewable-derived resource, lignin, were prepared by mixing ChCl and HBD at an appropriate molar ratio. • A fresh batch of the DES, ChCl–PCA, resulted in approximately 87% glucose yield, and the yield decreased to 78% and 70% for 2nd and 3rd runs, respectively. • Ease of synthesis, availability and economic viability of the components make DESs attractive and a potential compelling alternative to ILs. Kim et al. (2018) Green Chemistry , doi: 10.1039/C7GC03029K Background • DESs have been gaining attention as inexpensive alternatives to conventional ILs used in biomass pretreatment. • Lignin-derived compounds can serve as raw materials for DES synthesis and use in a biorefinery context. Approach • Ten phenolic compounds were tested as a hydrogen bond donor to synthesize novel DESs at varying molar ratios with ChCl. • After initial screening, four DESs were selected chosen and they were used for biomass pretreatment of switchgrass. • After pretreatment, the residual DESs were washed out to remove potential inhibitory effects on enzymatic hydrolysis and subsequent fermentation steps by phenolic compounds. Saccharification yield from the DES pretreated switchgrass (expressed as the percentage of the theoretical maximum based on the initial glucan and xylan contents in the starting biomass). HBA: hydroxybenzoic acid; CAT: catechol; VAN: vanillin; PCA: p-coumaric acid. Biorefinery concept using renewable DESs derived from lignin
- 8. Linking secondary metabolites to gene clusters in six diverse Aspergillus species Background • The genus of Aspergillus contains fungi that are relevant to plant and human pathology, food biotechnology, enzyme production, model organisms, and a selection of extremophiles. • Filamentous fungi produce a diverse range of secondary metabolites (SMs), including compounds that can serve as chemicals, pharmaceuticals and related bioproducts. • No comparative analysis across genomes for the identification of gene clusters that produce SMs has been conducted previously. Approach • Four diverse Aspergillus species (A. campestris, A. novofumigatus, A. ochraceoroseus, and A. steynii) have been whole-genome PacBio sequenced to provide genetic references in three Aspergillus sections and coupled with two existing genomes. • Thirteen Aspergillus genomes were analyzed with comparative genomics to determine phylogeny and genetic diversity, and link secondary metabolites of interest to specific fungi. Outcomes and Impacts • We have identified putative SM clusters for aflatoxin, chlorflavonin, and ochrindol in A. ochraceoroseus, A. campestris, and A. steynii, respectively, and novofumigatonin, ent-cycloechinulin, and epi-aszonalenins in A. novofumigatus. • Provides new insight into how biological, biochemical, and genomic information can be combined to identify genes involved in the biosynthesis of specific SMs that may be of interest and use as bioproducts. Kjærbølling et al. (2018) Proceedings of the National Academy of Sciences , doi: 10.1073/pnas.1715954115 (A) Overview of the SM gene clusters predicted in A. novofumigatus and their homologs in the reference species. (B) Overview of the SM gene clusters predicted in A. fumigatus and their homologs in the reference species. (C) A Venn diagram of the A. fumigatus and A. novofumigatus SM gene clusters. (D) The number and different types of SM gene clusters predicted in A. fumigatus and A. novofumigatus. DMATS, dimethylallyl tryptophan synthase; NRPS, nonribosomal peptide synthetase; PKS, polyketide synthase; TC, terpene cyclase.
- 9. Characterization of lignin streams during bionic liquid-based pretreatment Background • Cholinium lysinate ([Ch][Lys]), an example of a bionic liquid, is reported to have excellent biomass pretreatment efficiency and successfully employed in a one-pot IL-based configuration, with improved economics. • It is imperative to have a qualitative and quantitative understanding of the partitioning of lignin in the different process streams by performing a detailed mass balance analysis and structural characterization. Approach • A complete mass balance analysis of different components after [Ch][Lys] pretreatment followed by enzymatic saccharification of switchgrass, eucalyptus, and pine. • The lignin streams generated were characterized using a two-dimensional (2D) 1H–13C heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) spectroscopy, gel permeation chromatography (GPC), and gas chromatography–mass spectrometry (GC-MS). Outcomes and Impacts • Pretreatment of switchgrass resulted in a maximum reduction of lignin, whereas the lignin in pine remained primarily intact. • The 2D HSQC-NMR study, coupled with the SEC and compositional analysis data, revealed more carbohydrate residues after enzymatic hydrolysis for both eucalyptus and pine, indicating more recalcitrance, which will demand further process optimization. • Provides a deeper understanding of the origins of biomass recalcitrance as a function of genotype and pretreatment process. Dutta et al. (2018) ACS Sustainable Chemistry & Engineering , doi: 10.1021/acssuschemeng.7b02991 Mass balances (%DM) for the major biomass components of (A) switchgrass, (B) eucalyptus, and (C) pine during ionic liquid pre-treatment (IL-PT) with [Ch][Lys] and lignin streams (L1, L2L, L2S, L3L, L4S, L4L) generated during the process.