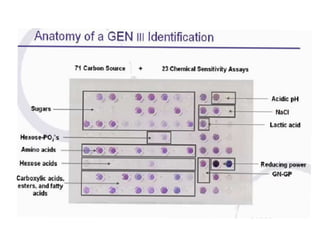
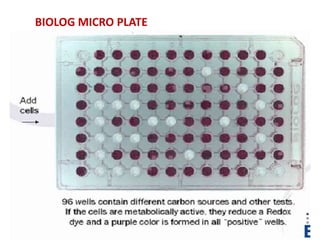
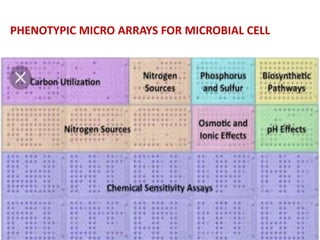
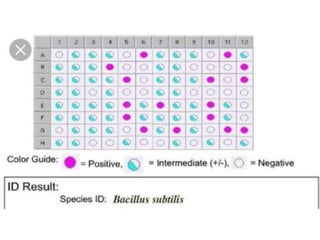
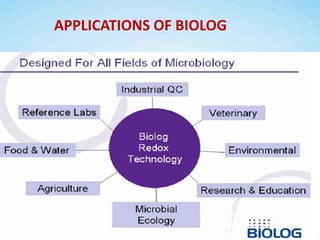
This document discusses the identification of bacteria using the Biolog system. It begins by explaining the importance of bacterial identification and limitations of traditional methods. It then describes how the Biolog system works, using a panel of 95 carbon sources to generate a metabolic fingerprint to identify bacteria. The procedure for using the Biolog system is outlined, including inoculating the microplates, incubating them, and obtaining readings to compare to the Biolog database for species-level identification. Key features of the Biolog system are its ease of use, ability to identify both gram-positive and gram-negative bacteria from a small number of cells, and generation of an identification within 24 hours.