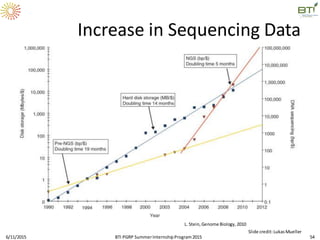
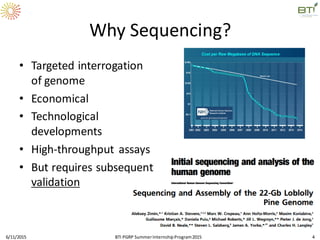
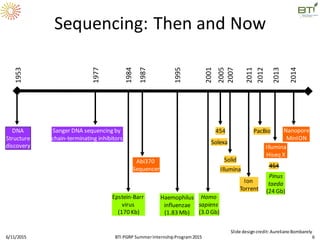
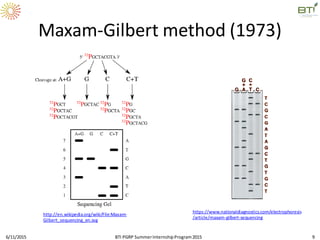
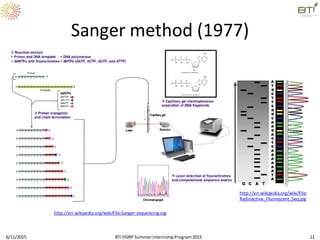
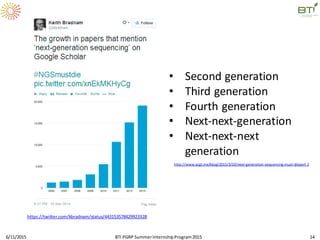
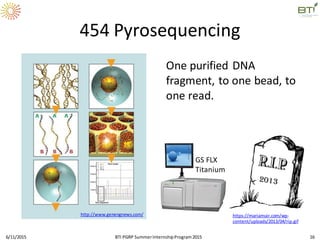
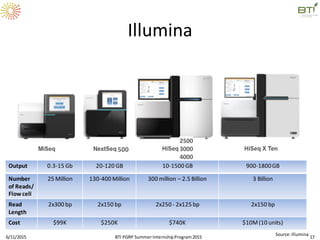
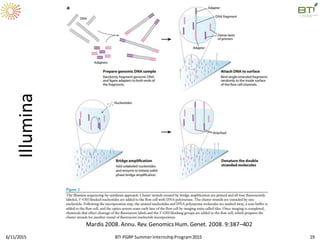
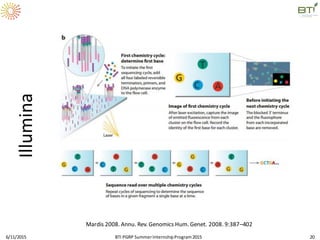
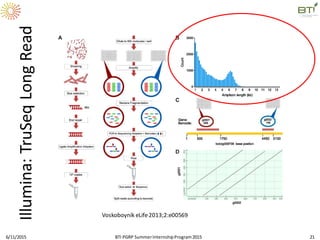
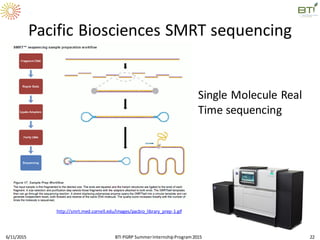
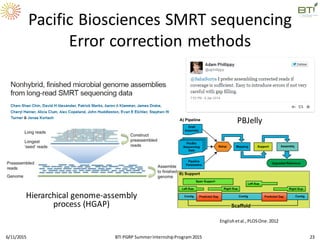
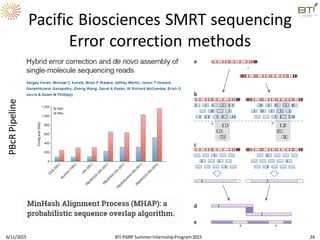
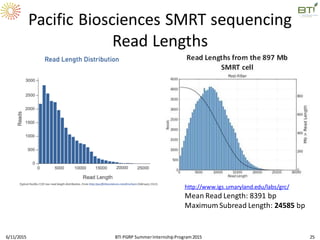
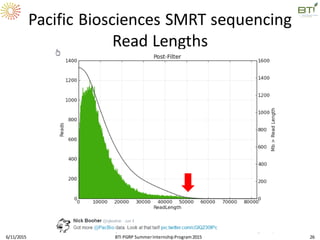
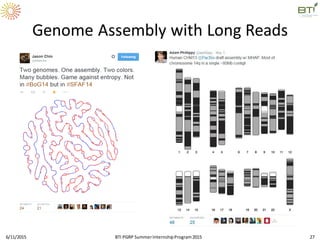
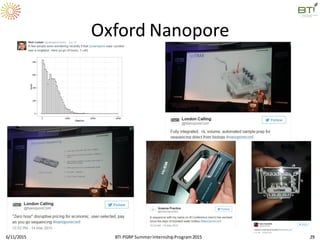
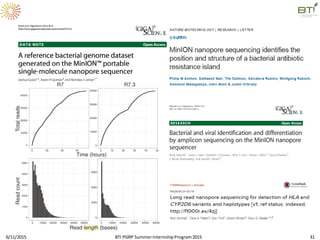
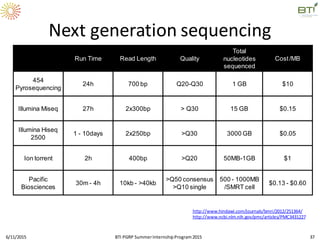
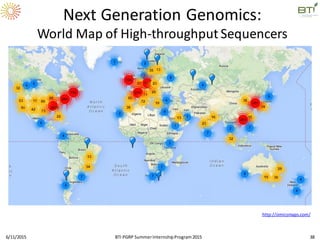
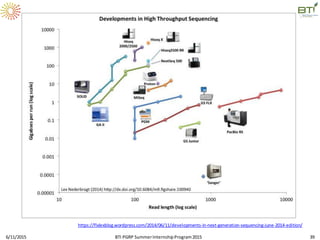
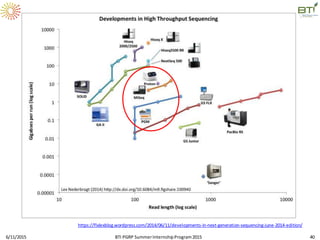
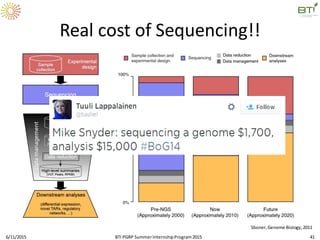
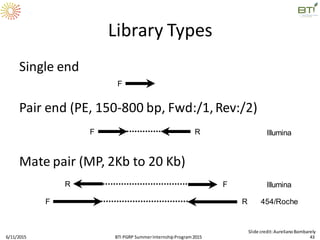
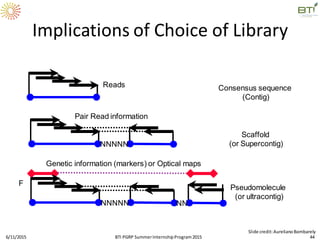
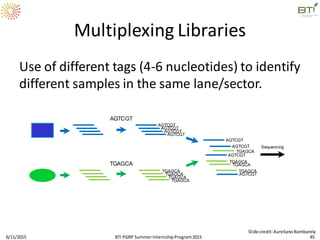
The document discusses advancements in genomic sequencing, including the evolution of sequencing technologies from Sanger to next-generation sequencing methods. It emphasizes the significance of various sequencing technologies, their output capabilities, cost-effectiveness, and the increasing volume of data generated. Additionally, it highlights the role of bioinformatics in processing and analyzing sequencing data.















![6/11/2015 48
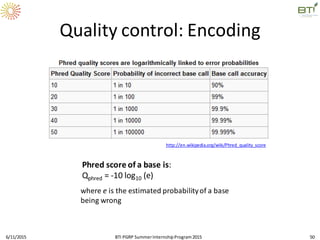
Quality control: Encoding
Fastq files:
!"#$%&'()*+,-./0123456789 Offset by 33 (Phred+33)
KLMNOPQRSTUVWXYZ[]^_`abcdefgh Offset by 64 (Phred+64)
BTI PGRP SummerInternshipProgram2015](https://image.slidesharecdn.com/sequencingpgrp2015-150611135124-lva1-app6891/85/Sequencing-and-Bioinformatics-PGRP-Summer-2015-48-320.jpg)
![Quality control: Encoding
6/11/2015 49
!"#$%&'()*+,-./0123456789 Offset by 33 (Phred+33)
KLMNOPQRSTUVWXYZ[]^_`abcdefgh Offset by 64 (Phred+64)
BTI PGRP SummerInternshipProgram2015](https://image.slidesharecdn.com/sequencingpgrp2015-150611135124-lva1-app6891/85/Sequencing-and-Bioinformatics-PGRP-Summer-2015-49-320.jpg)