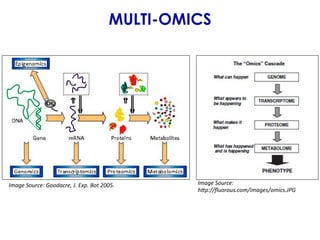
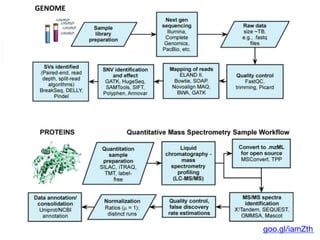
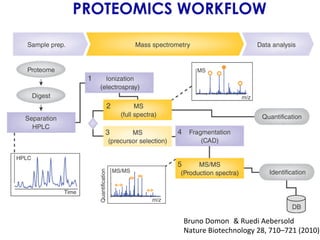
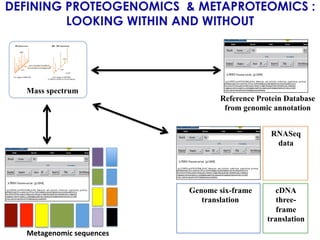
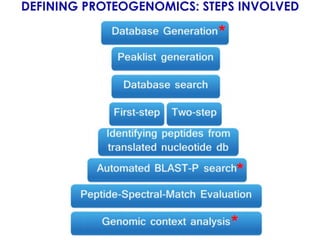
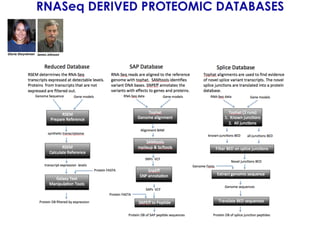
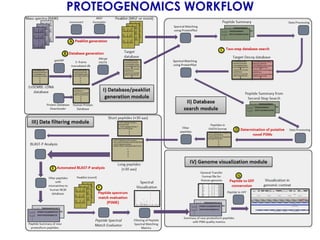
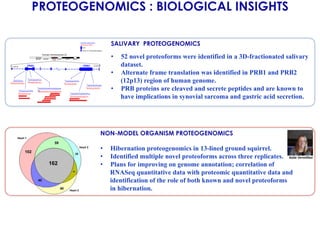
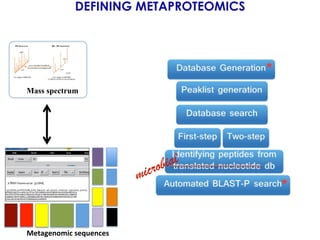
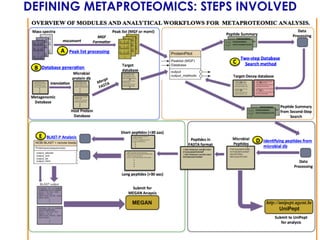
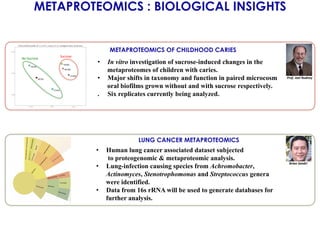
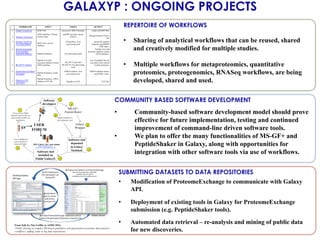
The document outlines the Galaxy framework as a comprehensive bioinformatics solution for multi-omic data analysis, including proteogenomics and metaproteomics. It highlights various ongoing research projects, such as identifying novel proteoforms in salivary and hibernation proteogenomics, and conducting metaproteomic analyses related to lung cancer and childhood caries. Additionally, it emphasizes community-based software development and the integration of workflows to enhance data analysis capabilities across various studies.