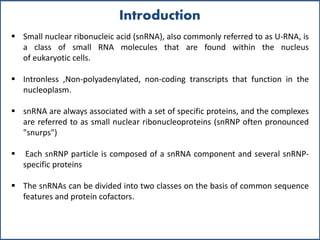
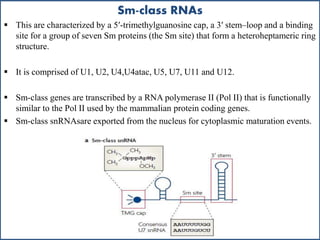
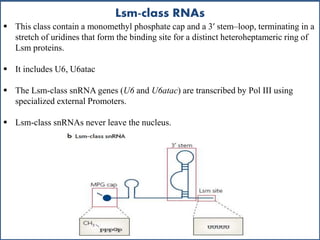
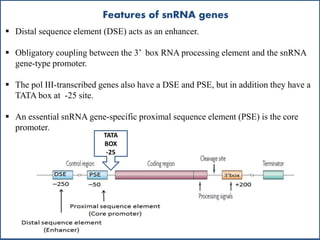
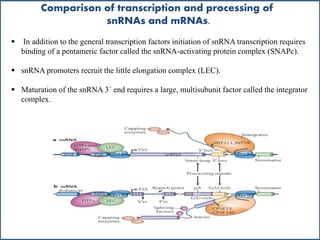
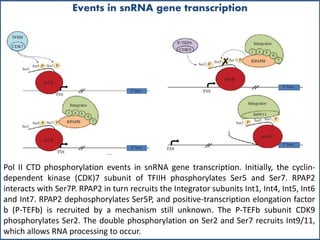
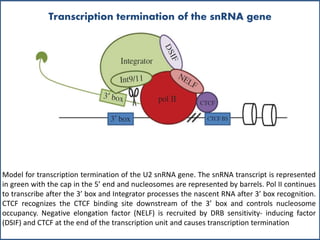
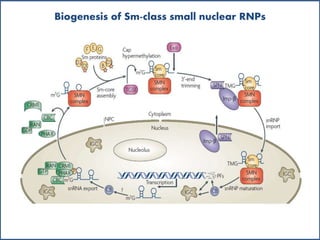
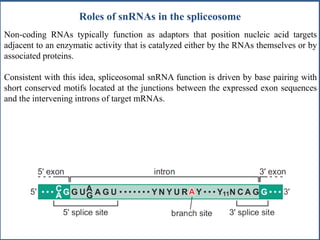
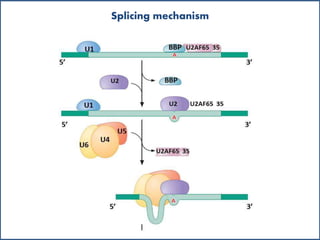
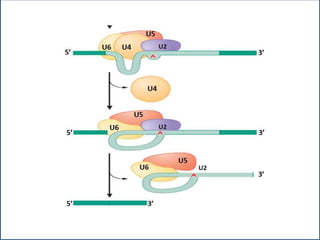
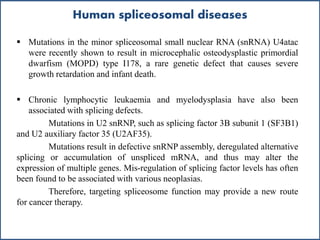
Small nuclear RNA (snRNA) is a class of non-coding RNA molecules found in the nucleus of eukaryotic cells, functioning primarily in RNA processing and splicing. It includes two classes, sm-class and lsm-class snRNAs, which have distinct structures and roles in gene transcription and RNA processing. The document also discusses the association of snRNAs with spliceosomal functions and the effects of mutations in snRNA related to various diseases.