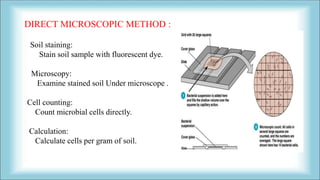
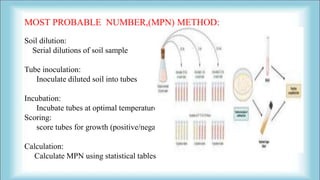
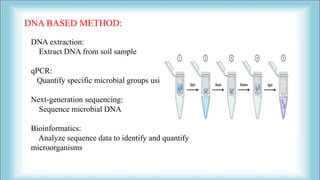
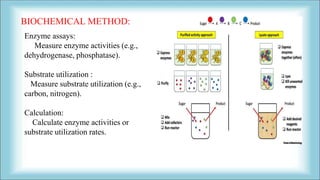
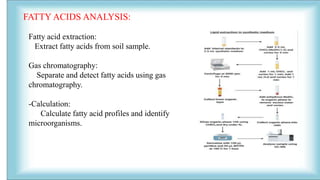
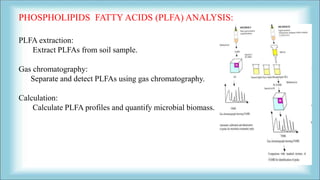
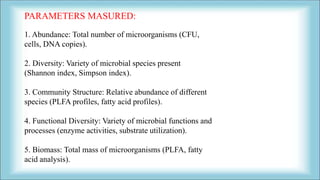
The document focuses on the quantification of soil microflora, which encompasses the measurement of microorganisms such as bacteria, fungi, and viruses in soil. It details various methods for quantification, including cultivation-based, molecular biology-based, and biochemical methods, as well as the principles and calculations involved in each. Additionally, it discusses the applications and limitations of quantifying soil microflora, emphasizing the challenges related to soil heterogeneity and methodological biases.