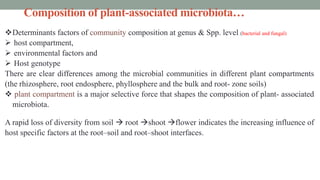
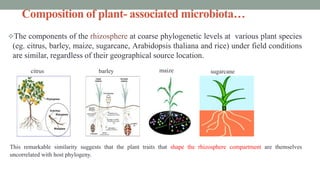
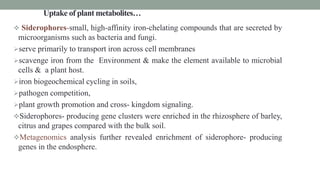
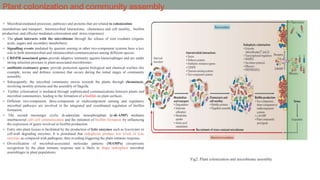
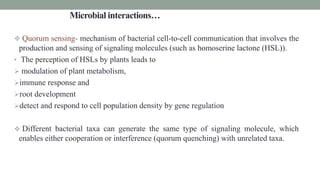
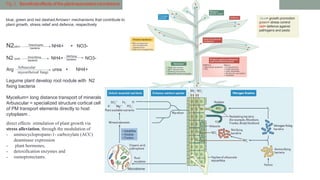
The document summarizes the plant microbiome, including its composition, core microbiota, and dynamics over time. It discusses how microorganisms colonize plants by sensing signals, uptaking metabolites, and evading defenses. Microbial interactions and functions like nutrient acquisition, disease resistance, and stress tolerance are also covered. While research has made progress understanding bacterial and fungal communities, gaps remain regarding other microorganisms and incorporating the microbiome into plant breeding.