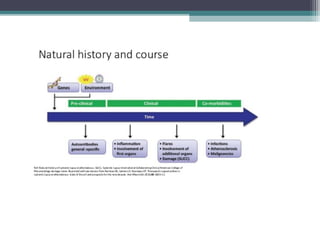
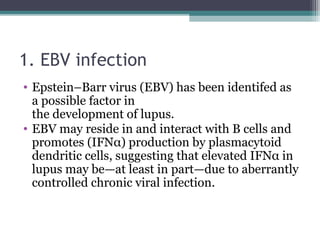
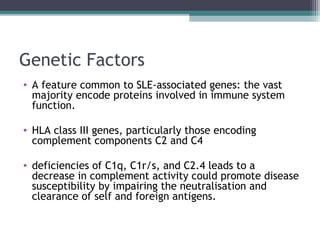
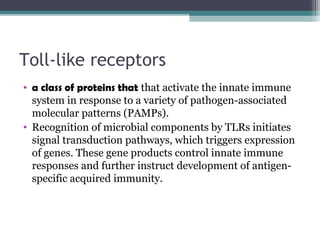
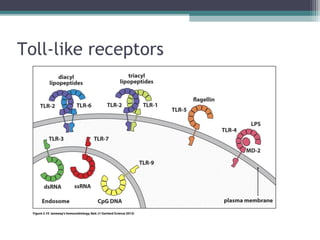
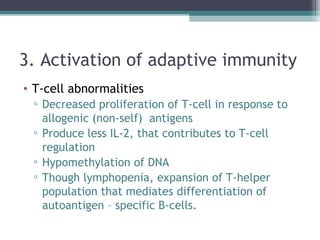
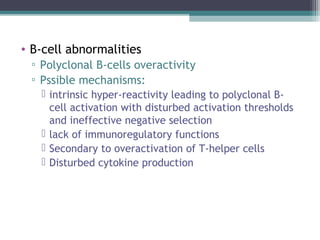
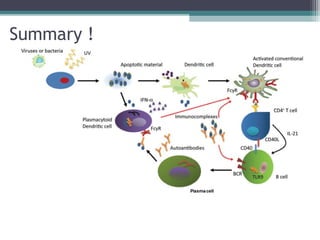
Systemic lupus erythematosus (SLE) is characterized by recurrent immune system activation leading to tissue damage, influenced by environmental, genetic, and hormonal factors. Key contributors include EBV infections, drug-induced lupus, and UV light, alongside a genetic predisposition with specific gene associations. Critical events in SLE pathogenesis involve apoptosis, abnormal nucleic acid recognition, and dysfunctional innate and adaptive immune responses, resulting in autoantibody production and tissue damage.