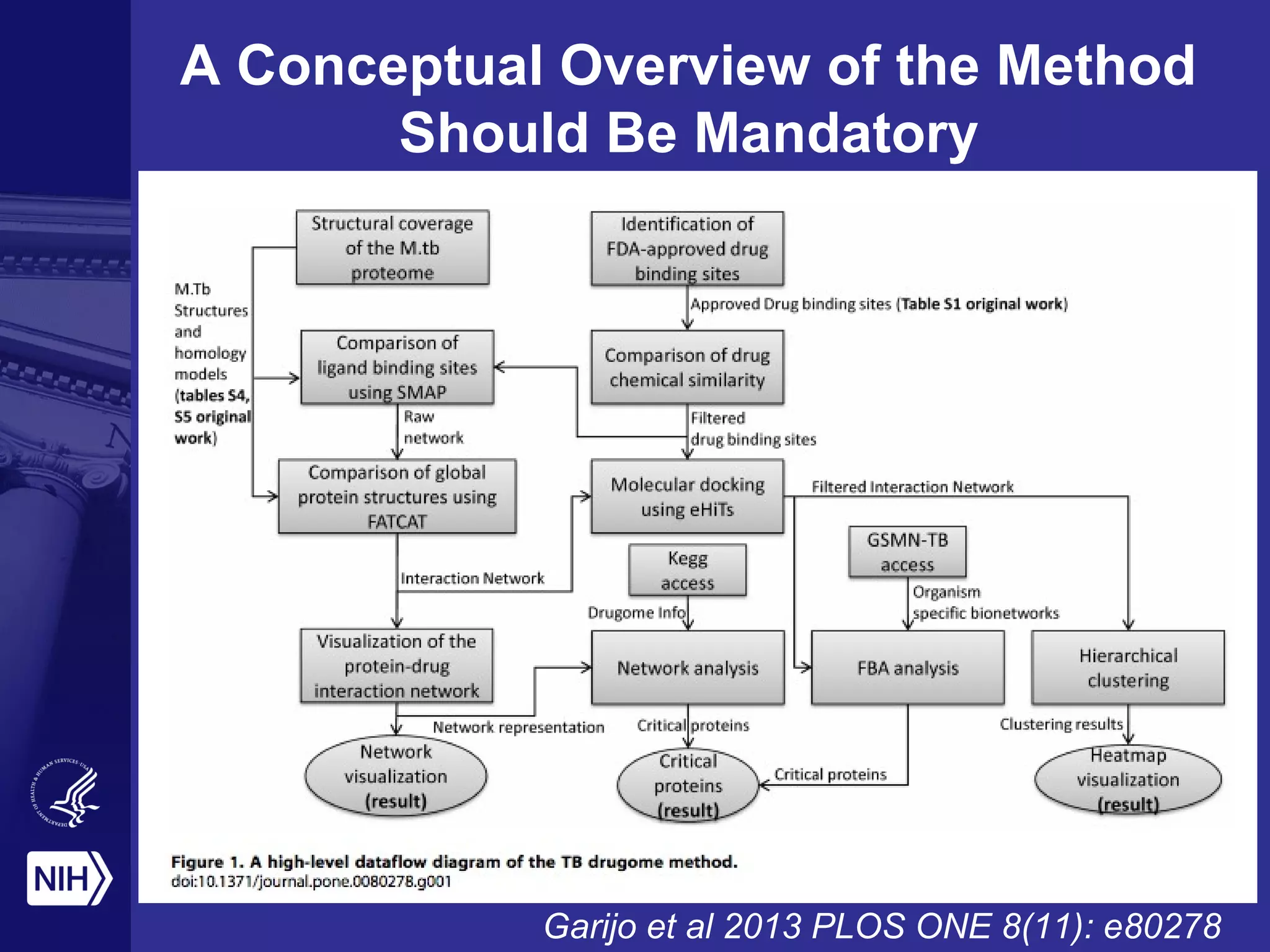
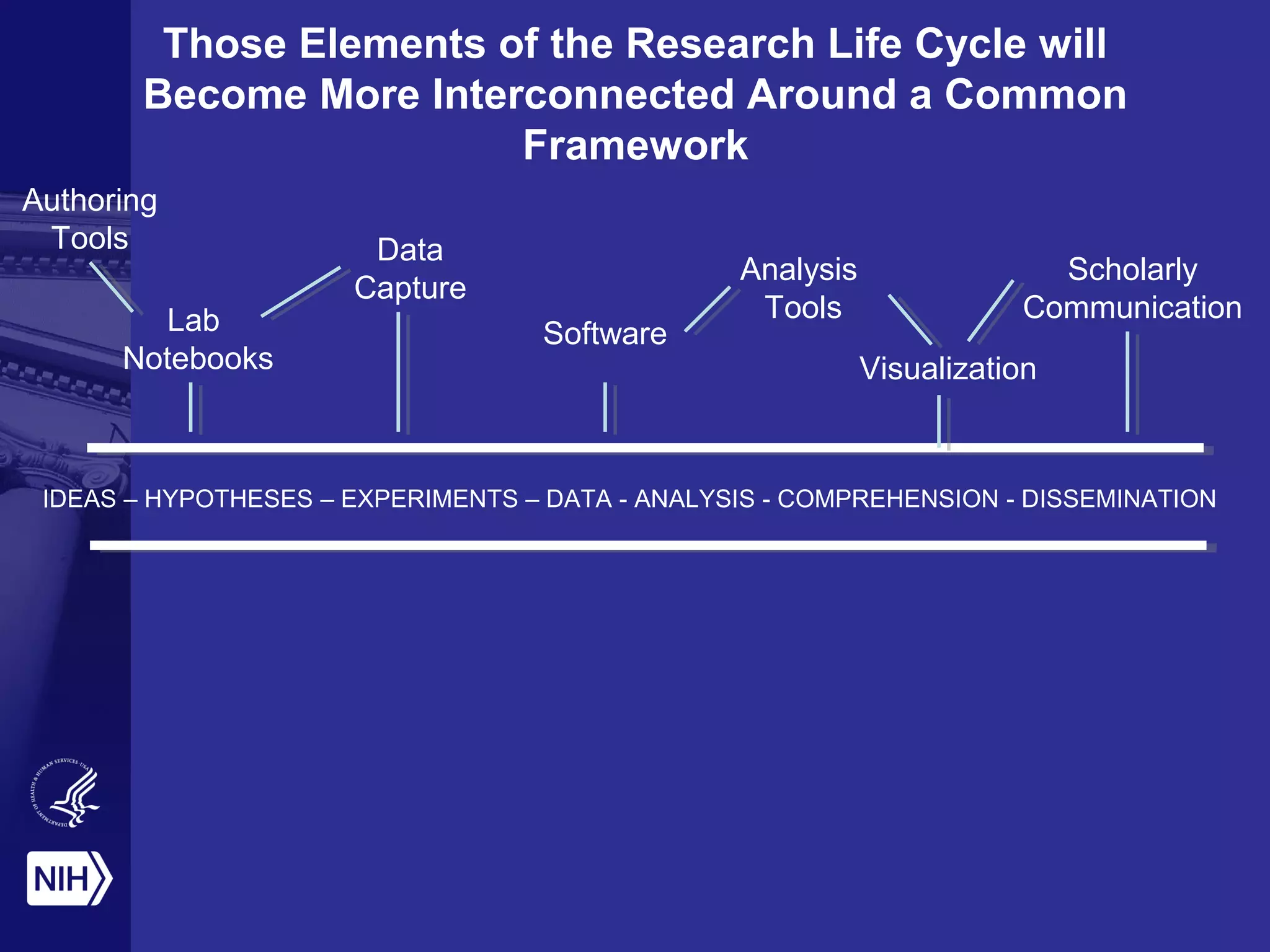
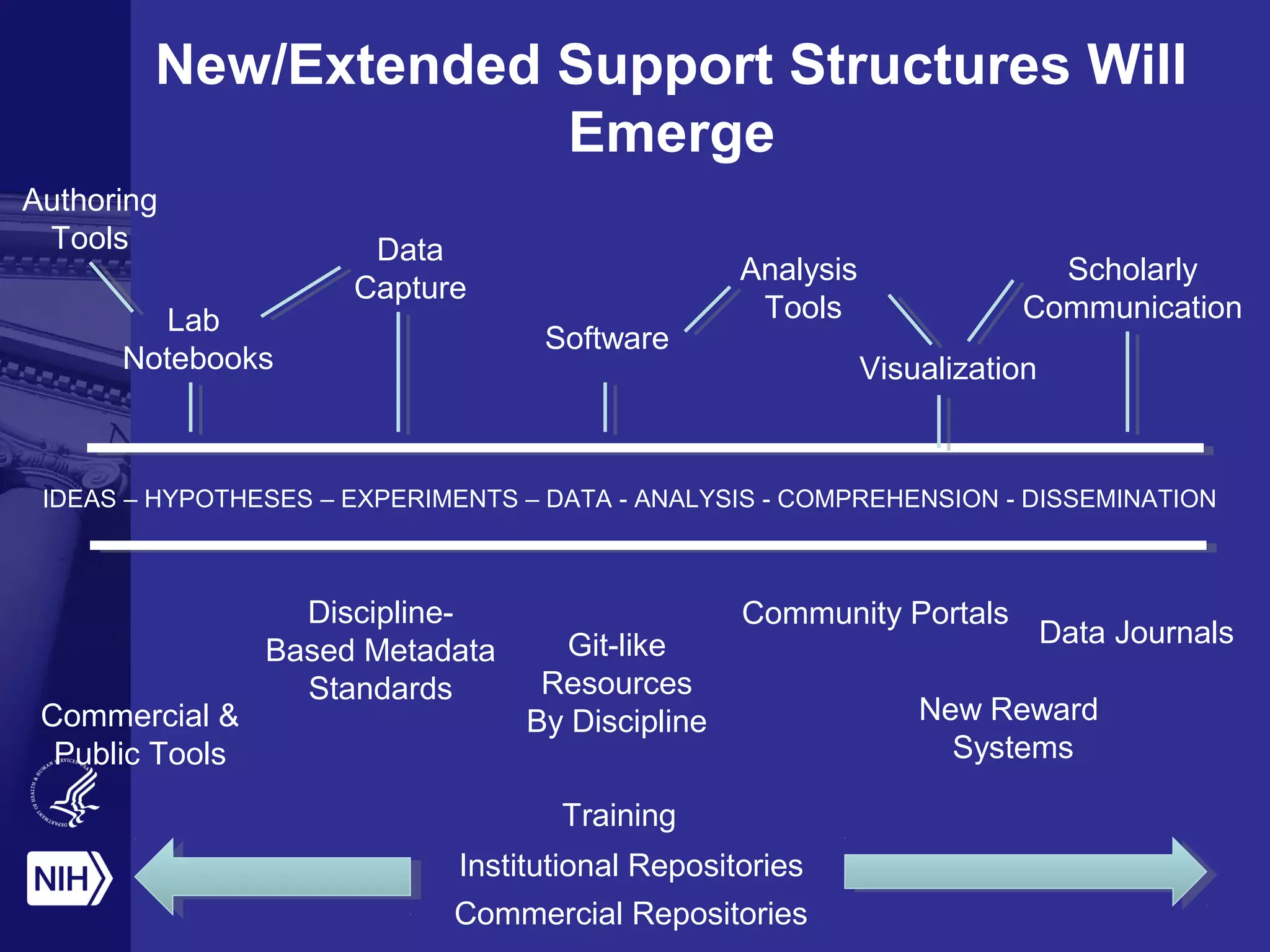
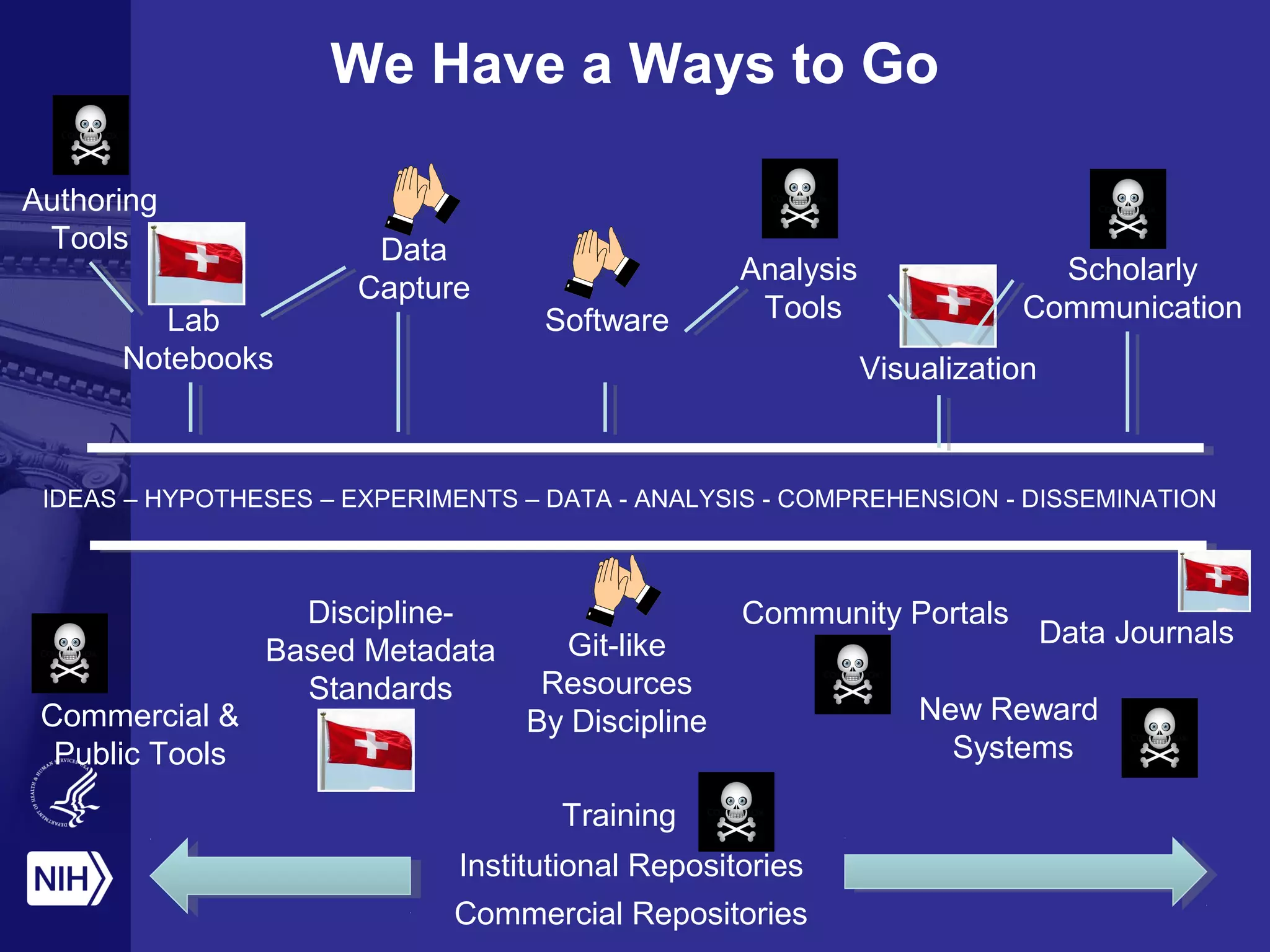
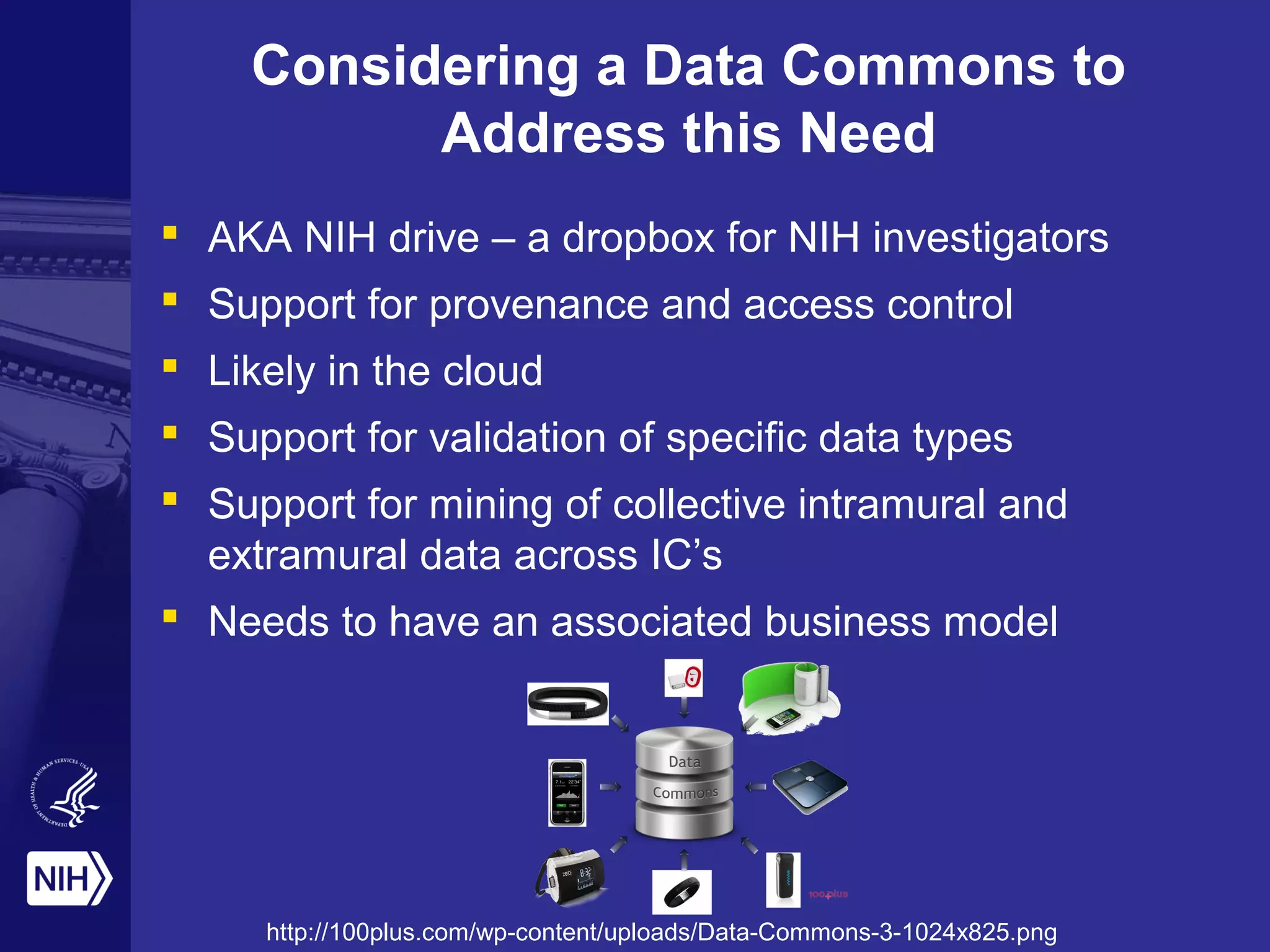
This document provides an overview of Philip Bourne's early observations and thoughts regarding data management at the NIH. Some of the key points are: 1) Existing data resources are not well understood in terms of how they are used; 2) There is a need to focus on how data will be managed and shared, not just why it should be; 3) There is no NIH-wide sustainability plan for data management; 4) Training in biomedical data science is inconsistent. The document discusses some potential solutions such as establishing a NIH data commons and improving training programs.



![Numberofreleasedentries
Year
Motivation for Change:
PDB Growth in Numbers and Complexity
[From the RCSB Protein Data Bank]](https://image.slidesharecdn.com/ontologynsf042814-140428105338-phpapp01/75/Data-at-the-NIH-Some-Early-Thoughts-4-2048.jpg)







![* http://www.cdc.gov/h1n1flu/estimates/April_March_13.htm
Jan. 2008 Jan. 2009 Jan. 2010Jul. 2009Jul. 2008 Jul. 2010
1RUZ: 1918 H1 Hemagglutinin
Structure Summary page activity for
H1N1 Influenza related structures
3B7E: Neuraminidase of A/Brevig Mission/1/1918
H1N1 strain in complex with zanamivir
[Andreas Prlic]
Consider What
Might Be Possible](https://image.slidesharecdn.com/ontologynsf042814-140428105338-phpapp01/75/Data-at-the-NIH-Some-Early-Thoughts-14-2048.jpg)









![47/53 “landmark” publications
could not be replicated
[Begley, Ellis Nature,
483, 2012] [Carole Goble]](https://image.slidesharecdn.com/ontologynsf042814-140428105338-phpapp01/75/Data-at-the-NIH-Some-Early-Thoughts-25-2048.jpg)