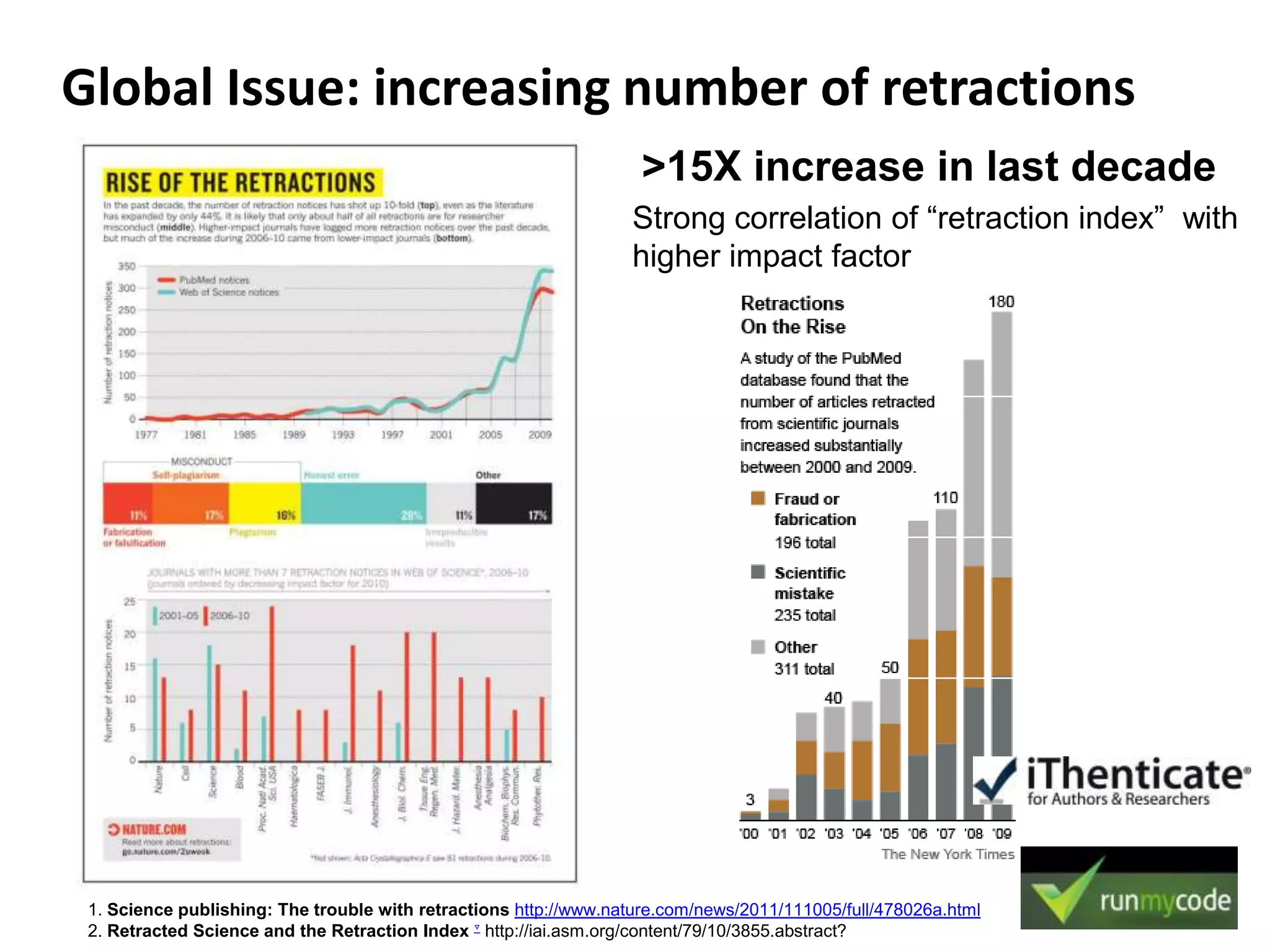
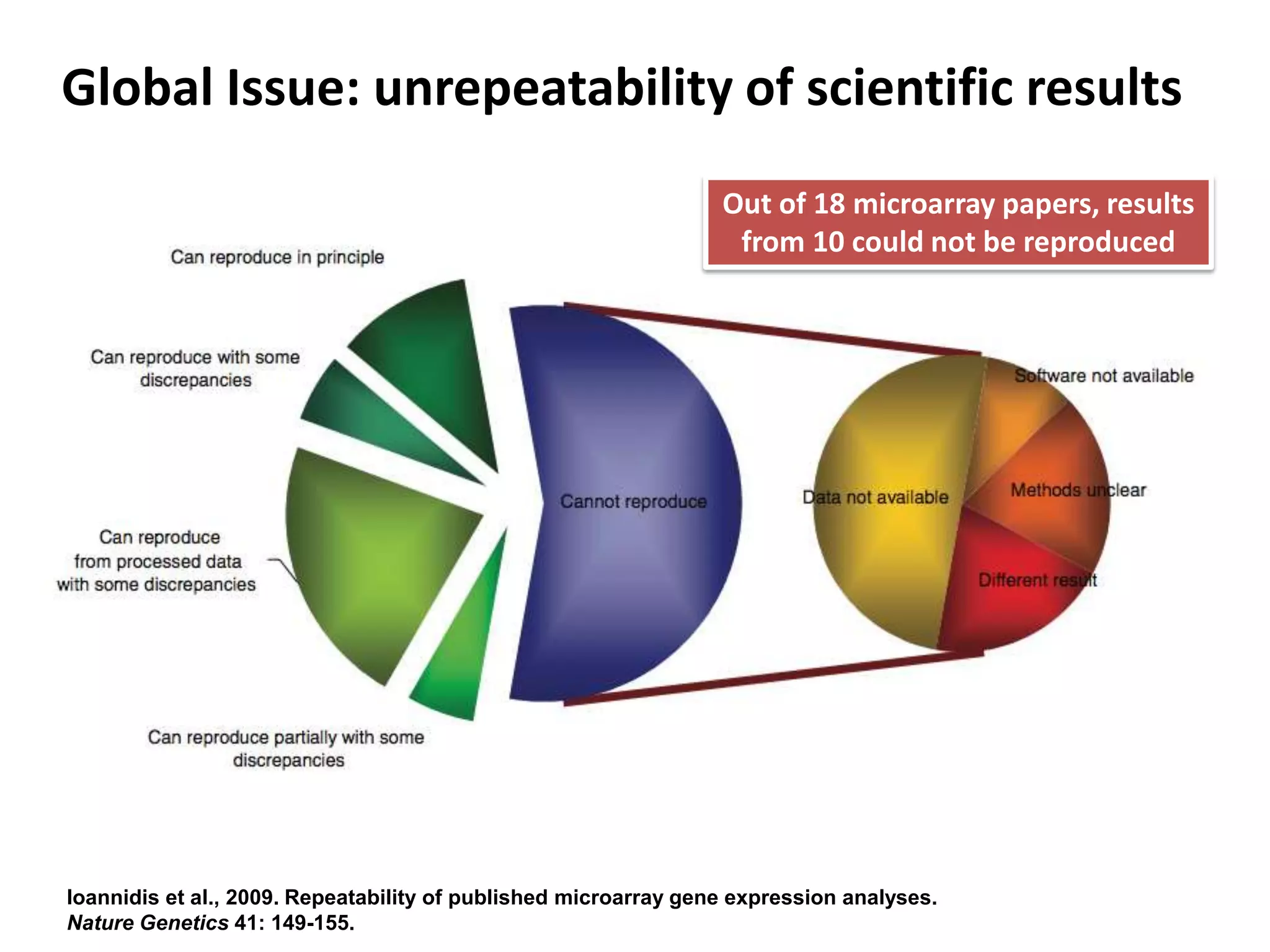
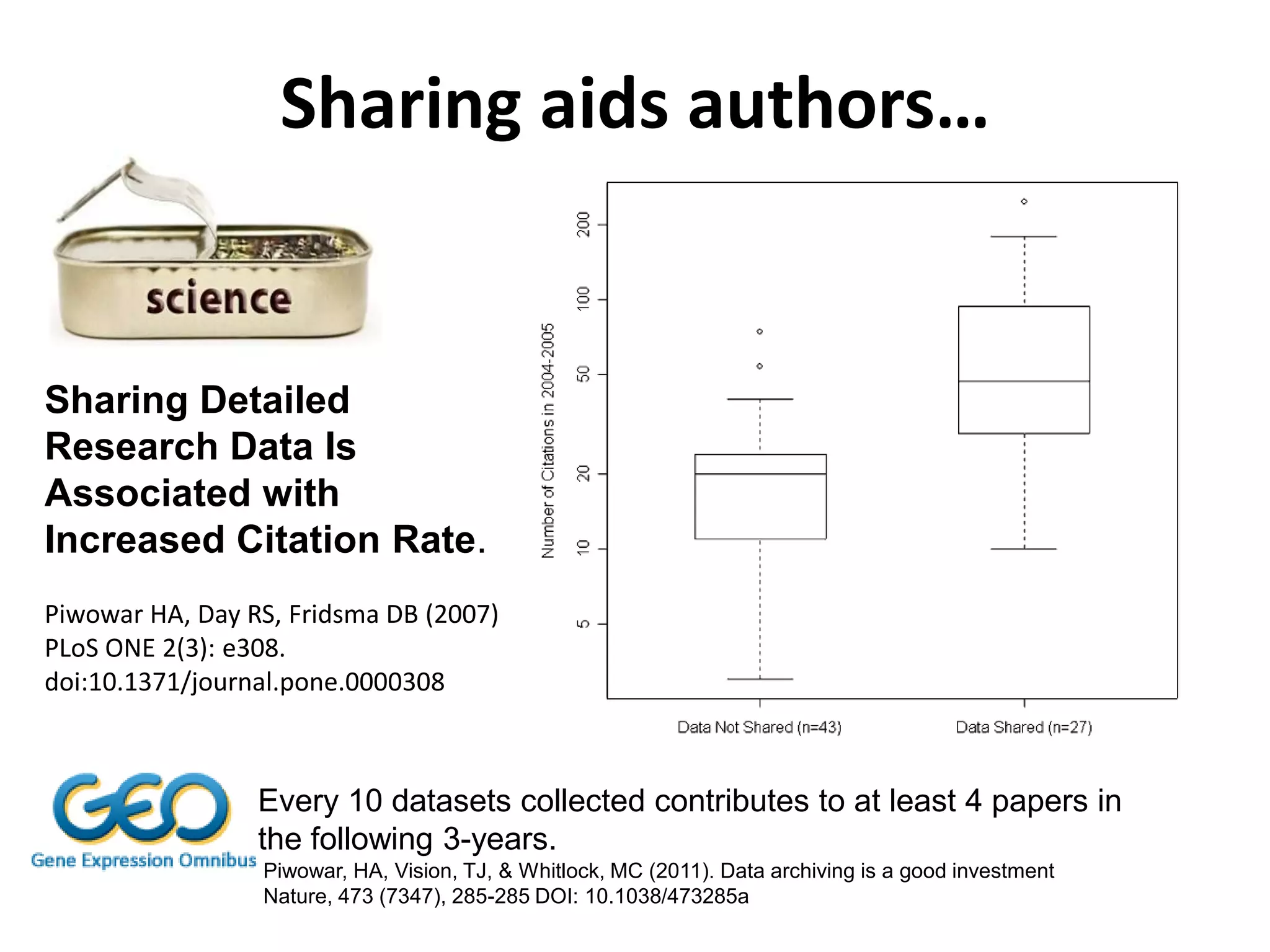
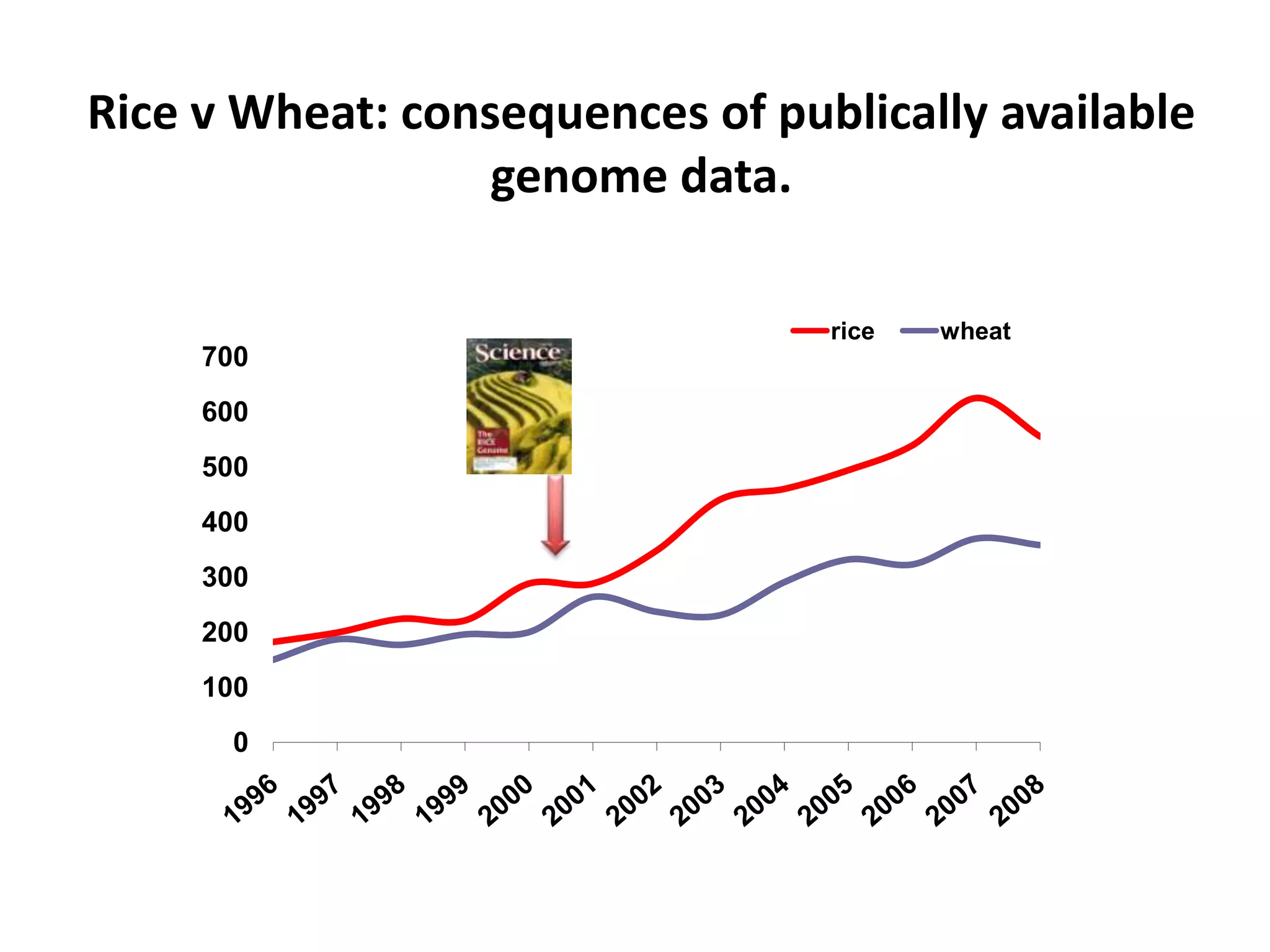
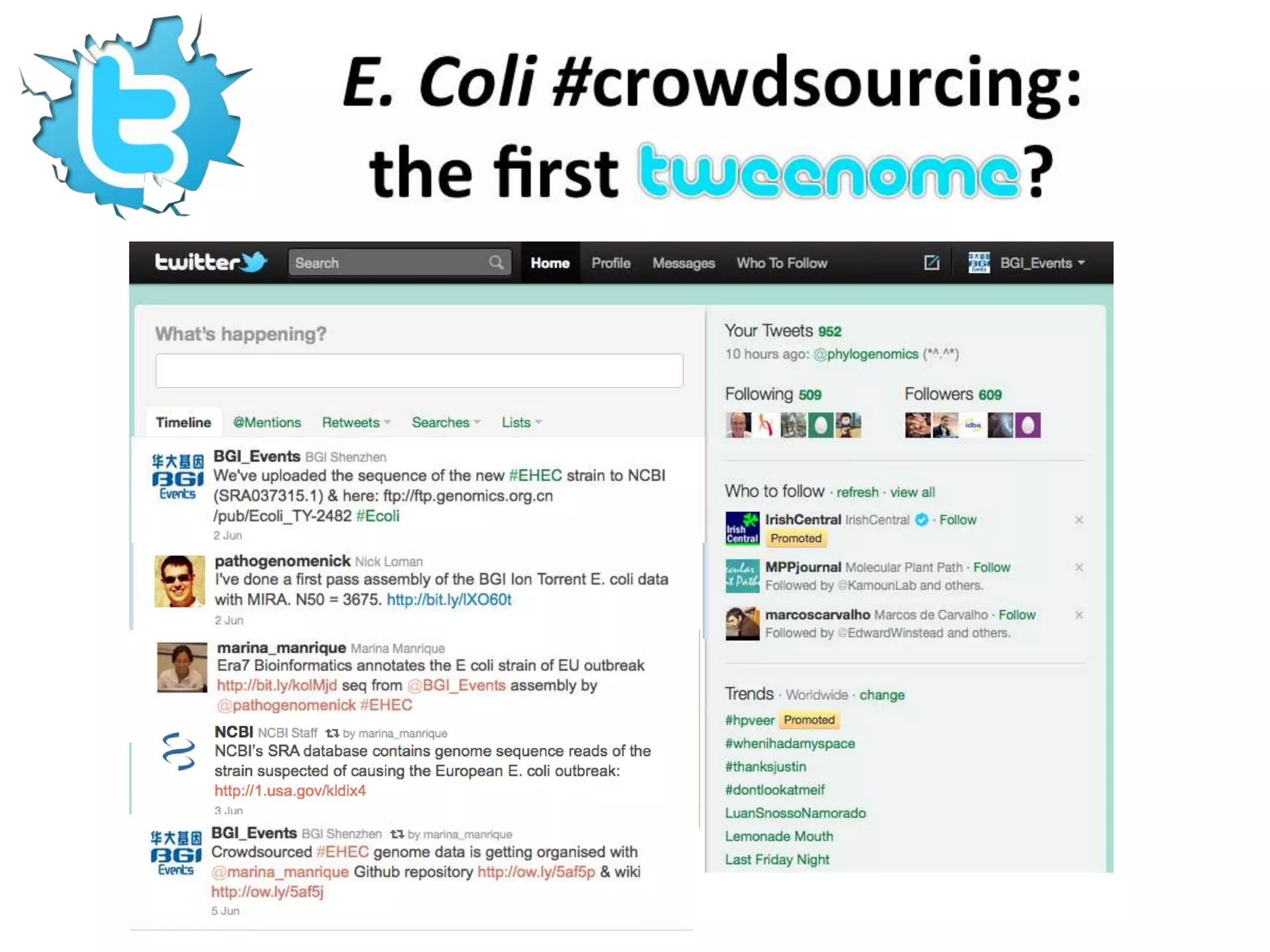
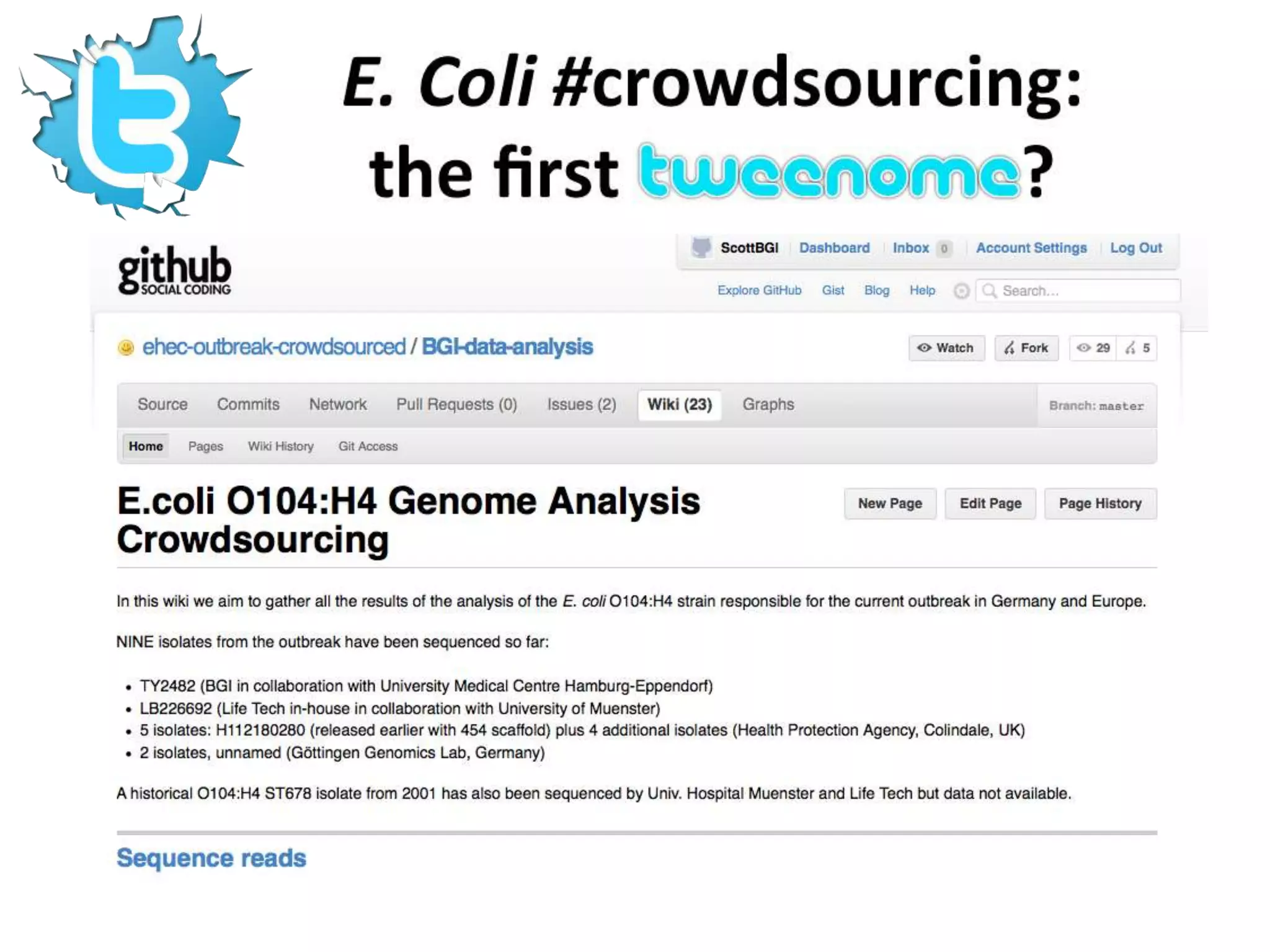
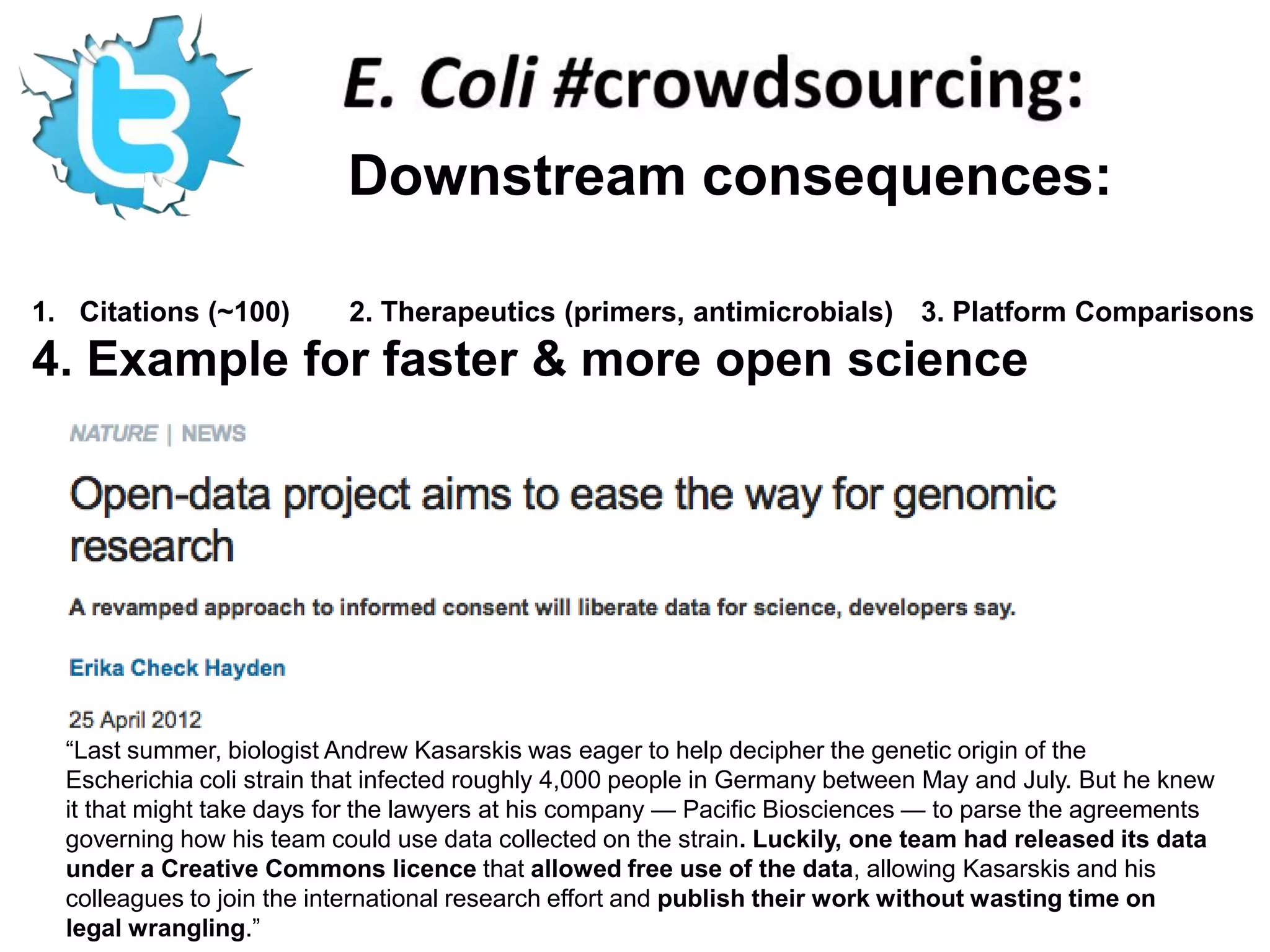
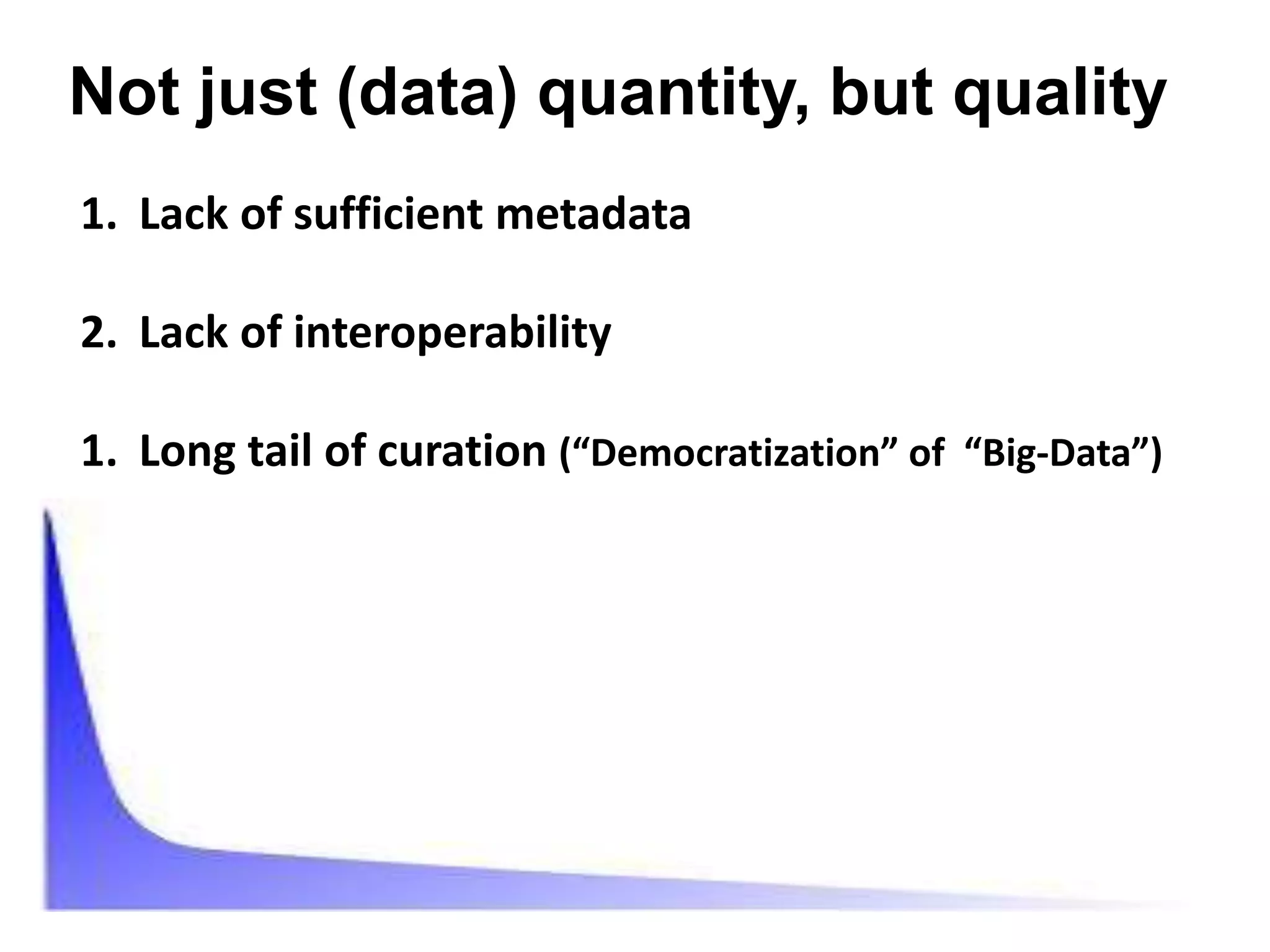
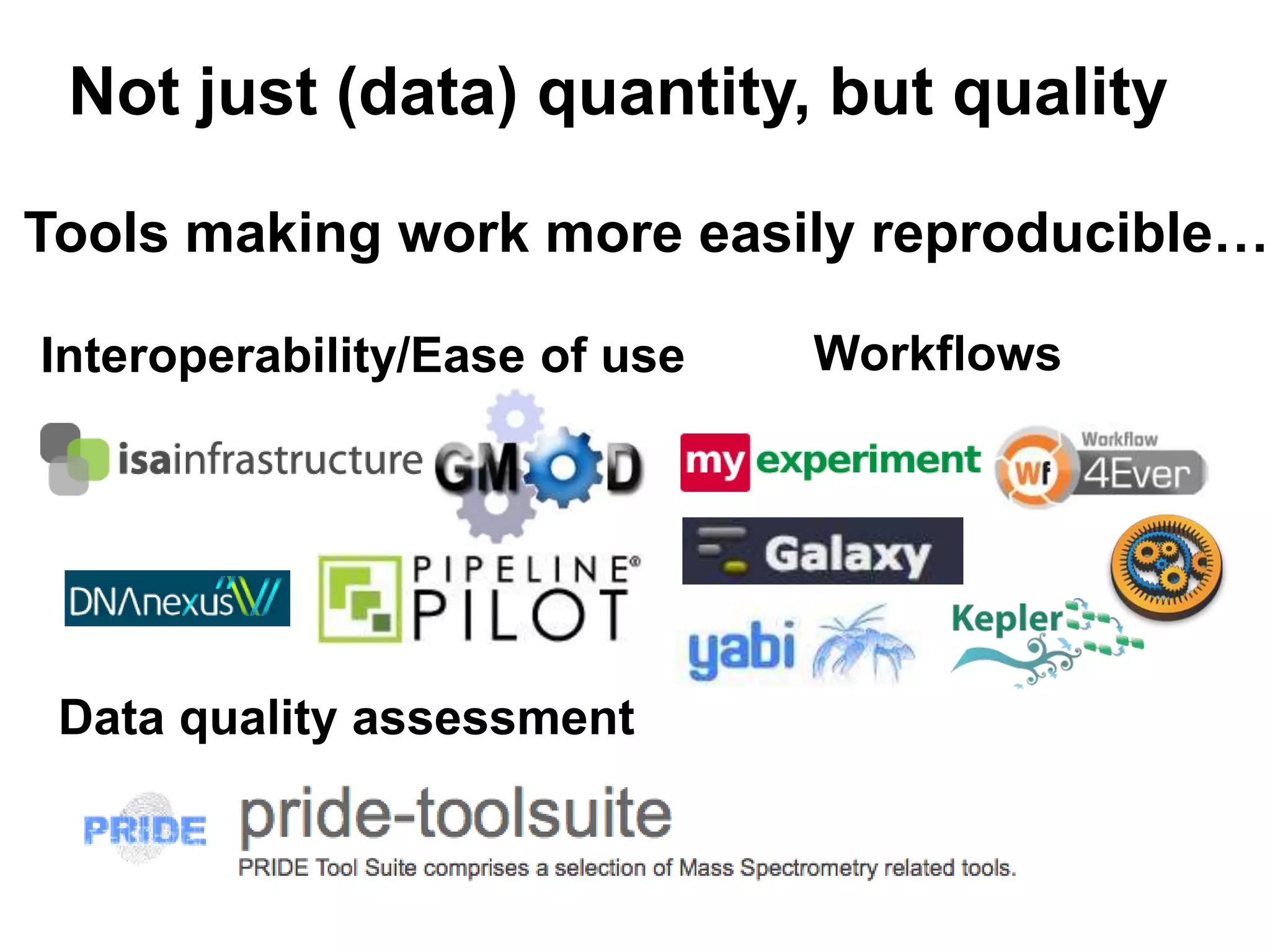
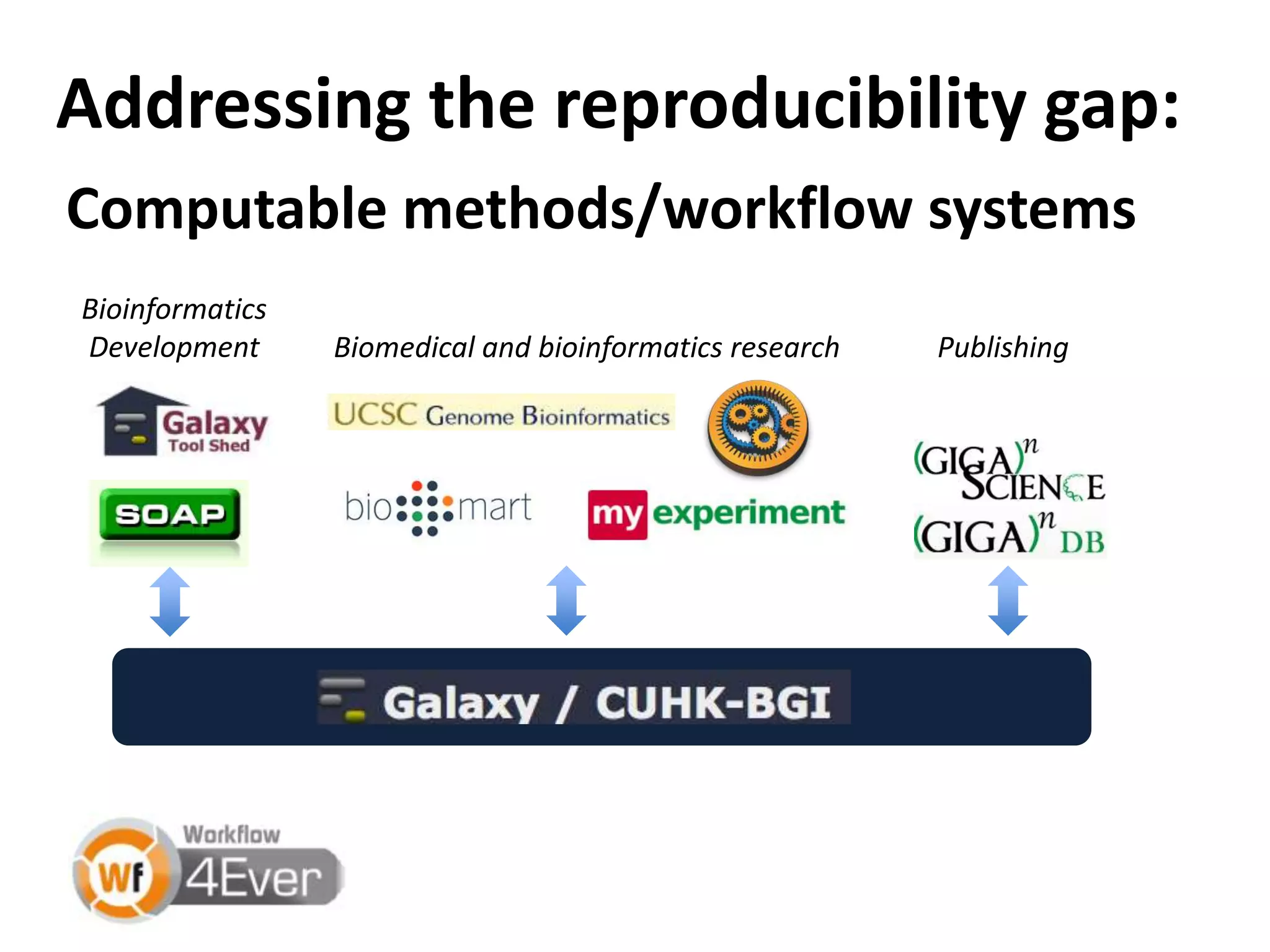
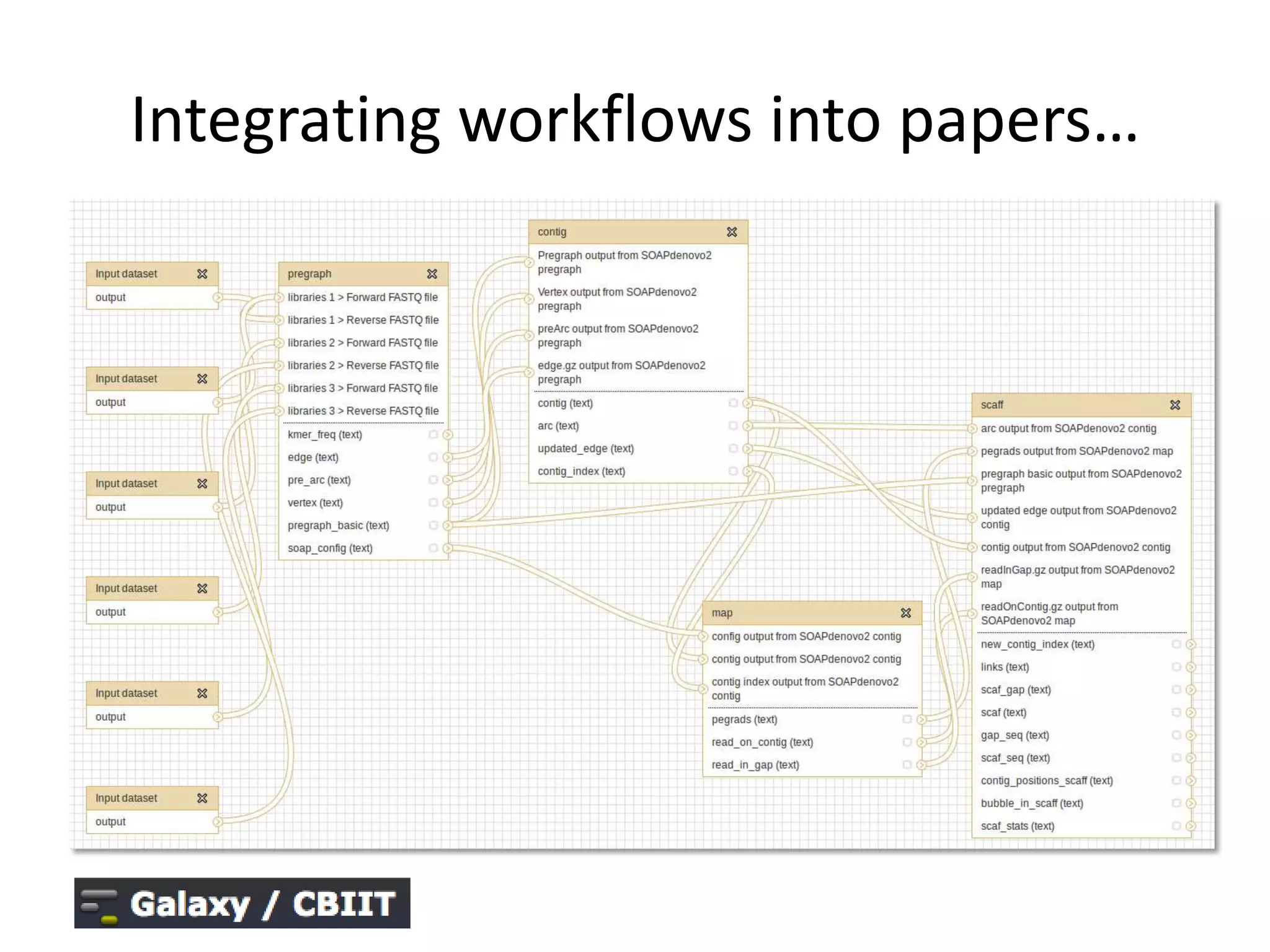
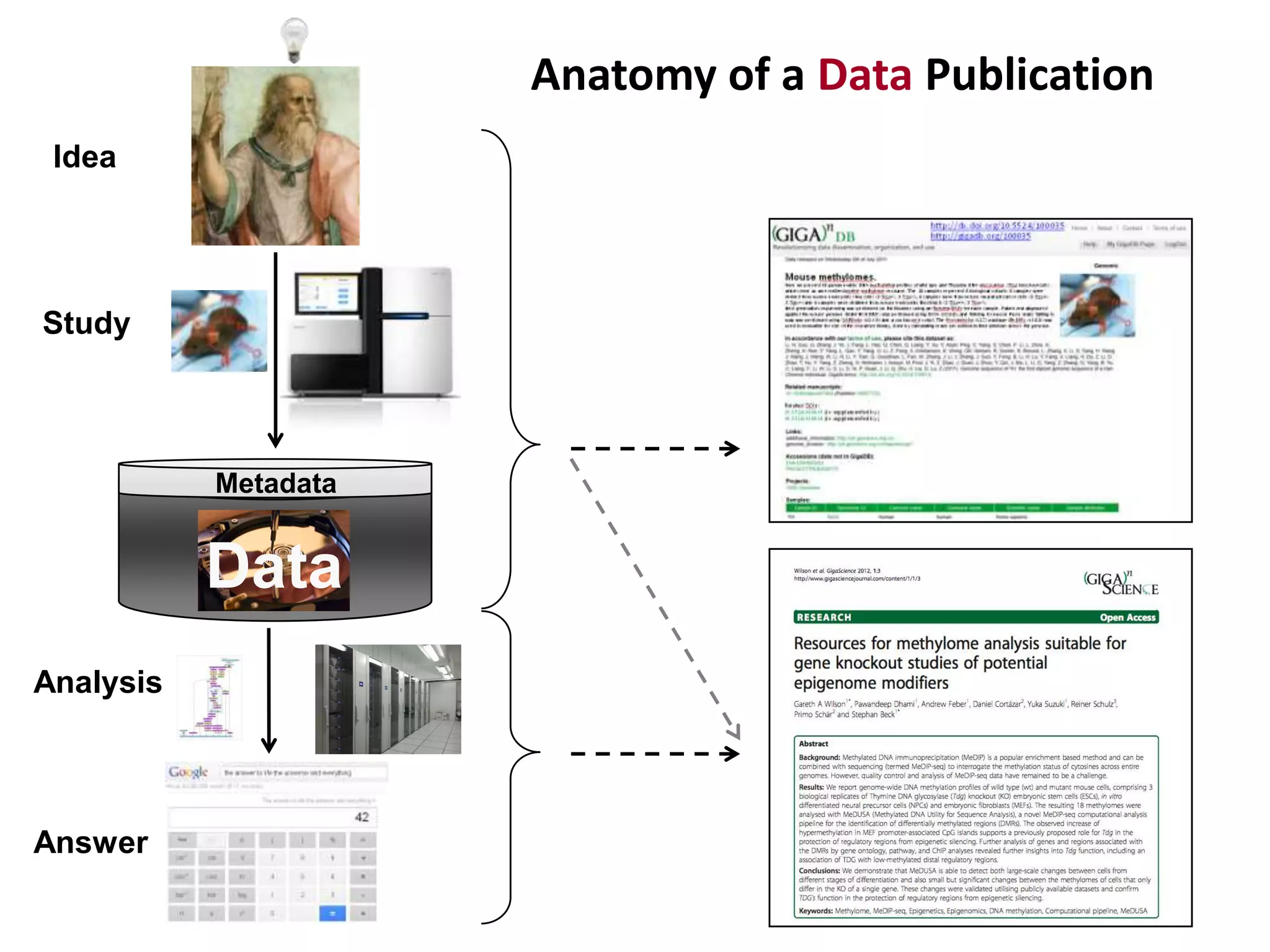
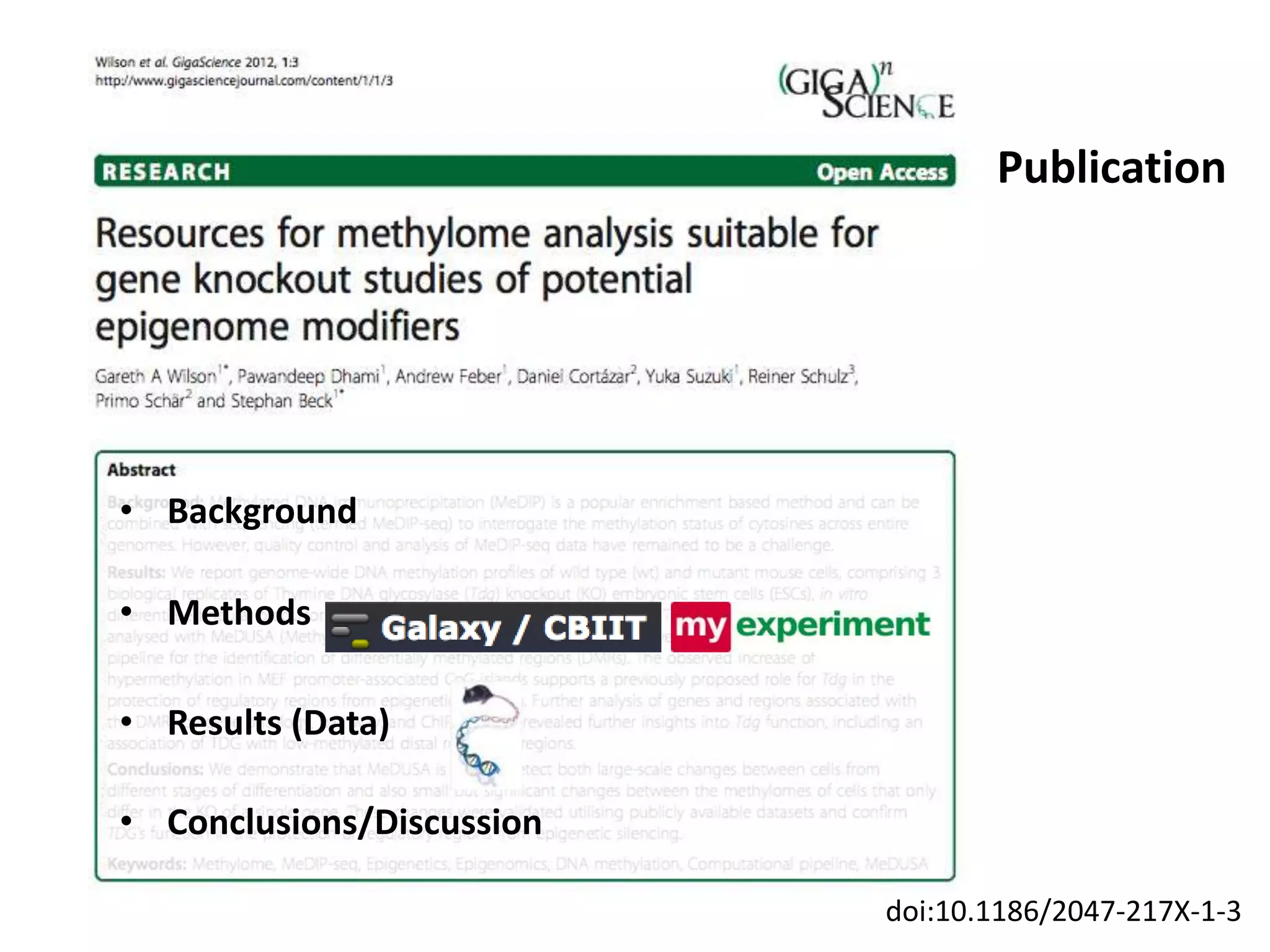
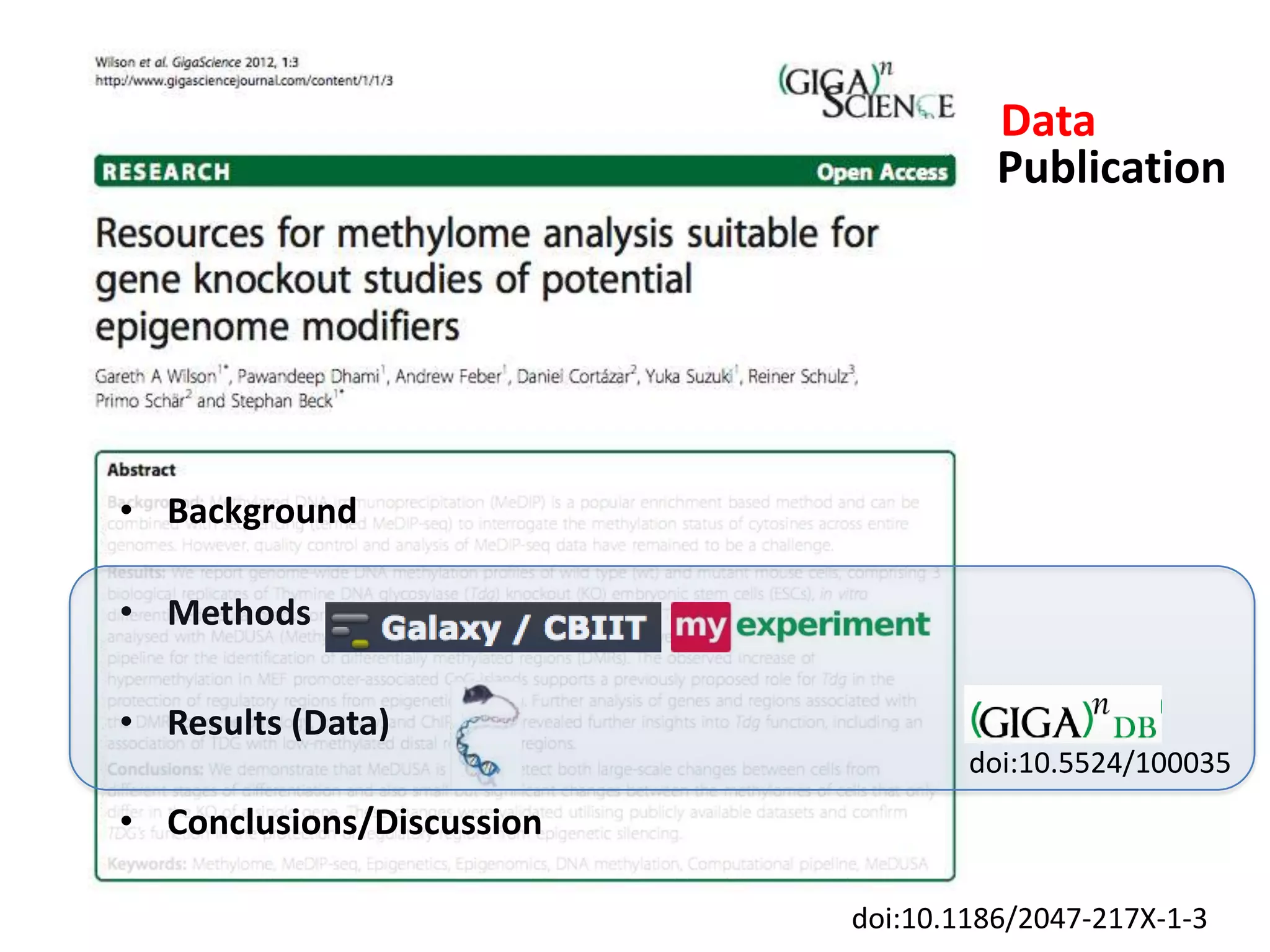
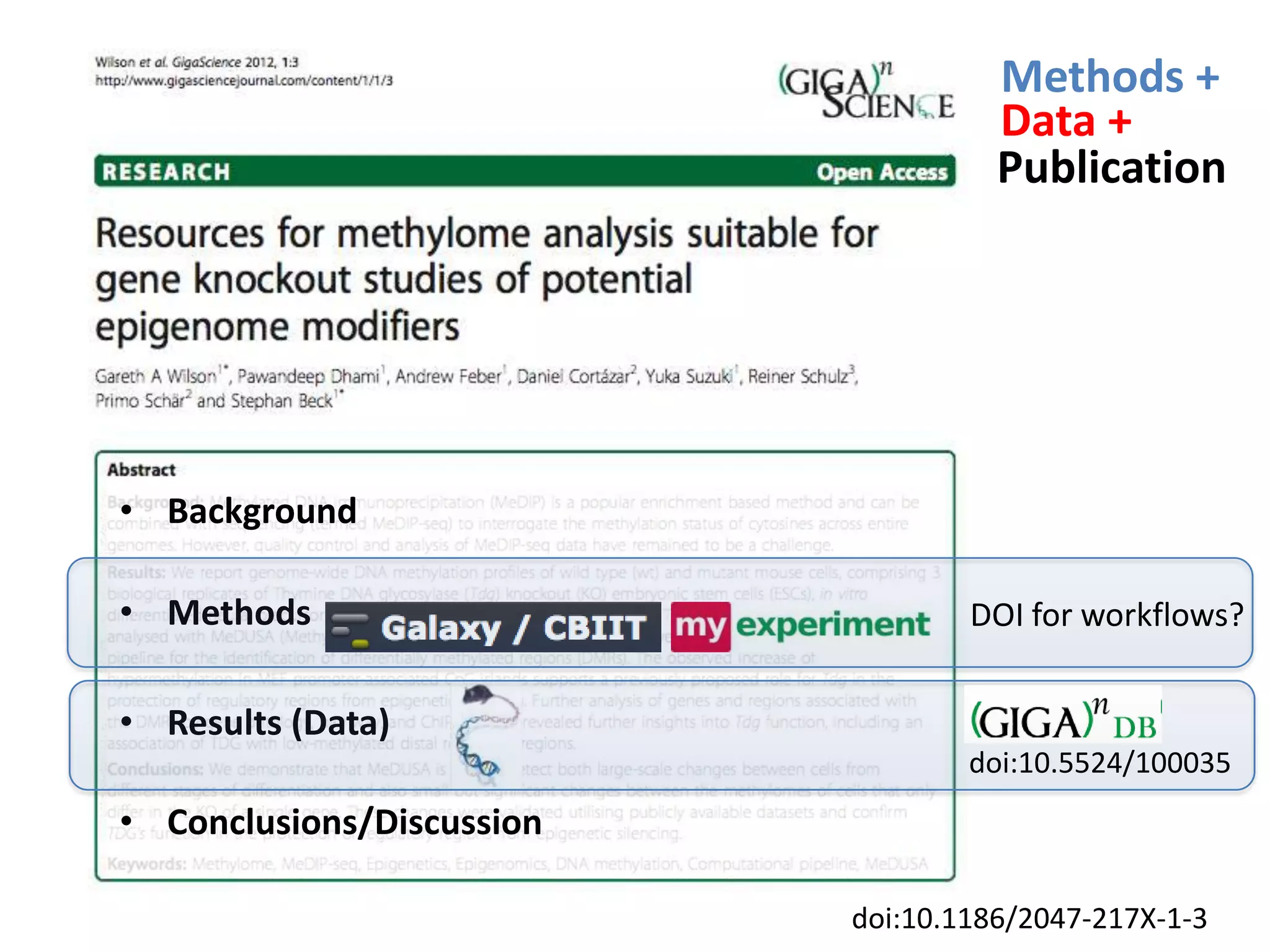
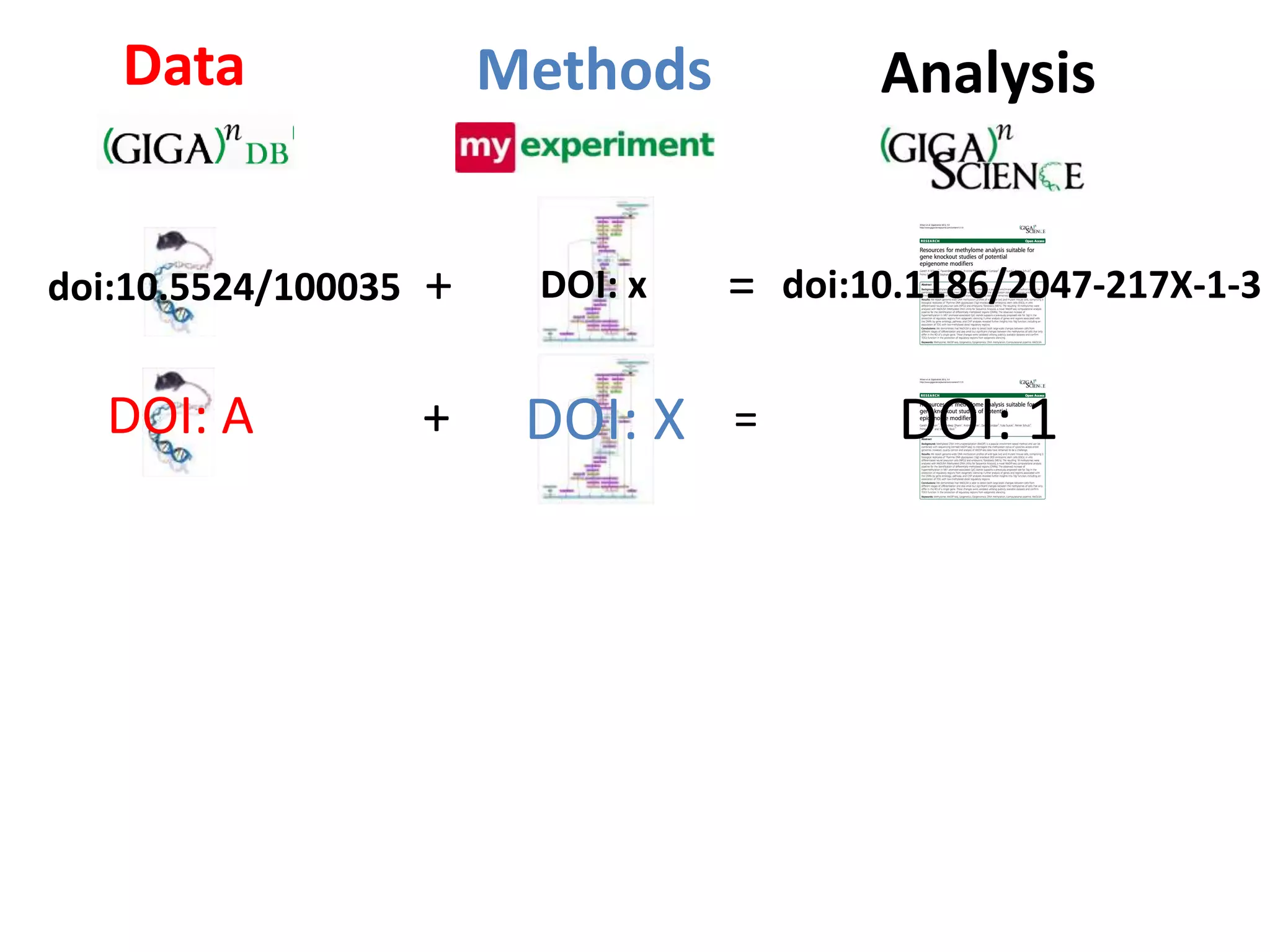
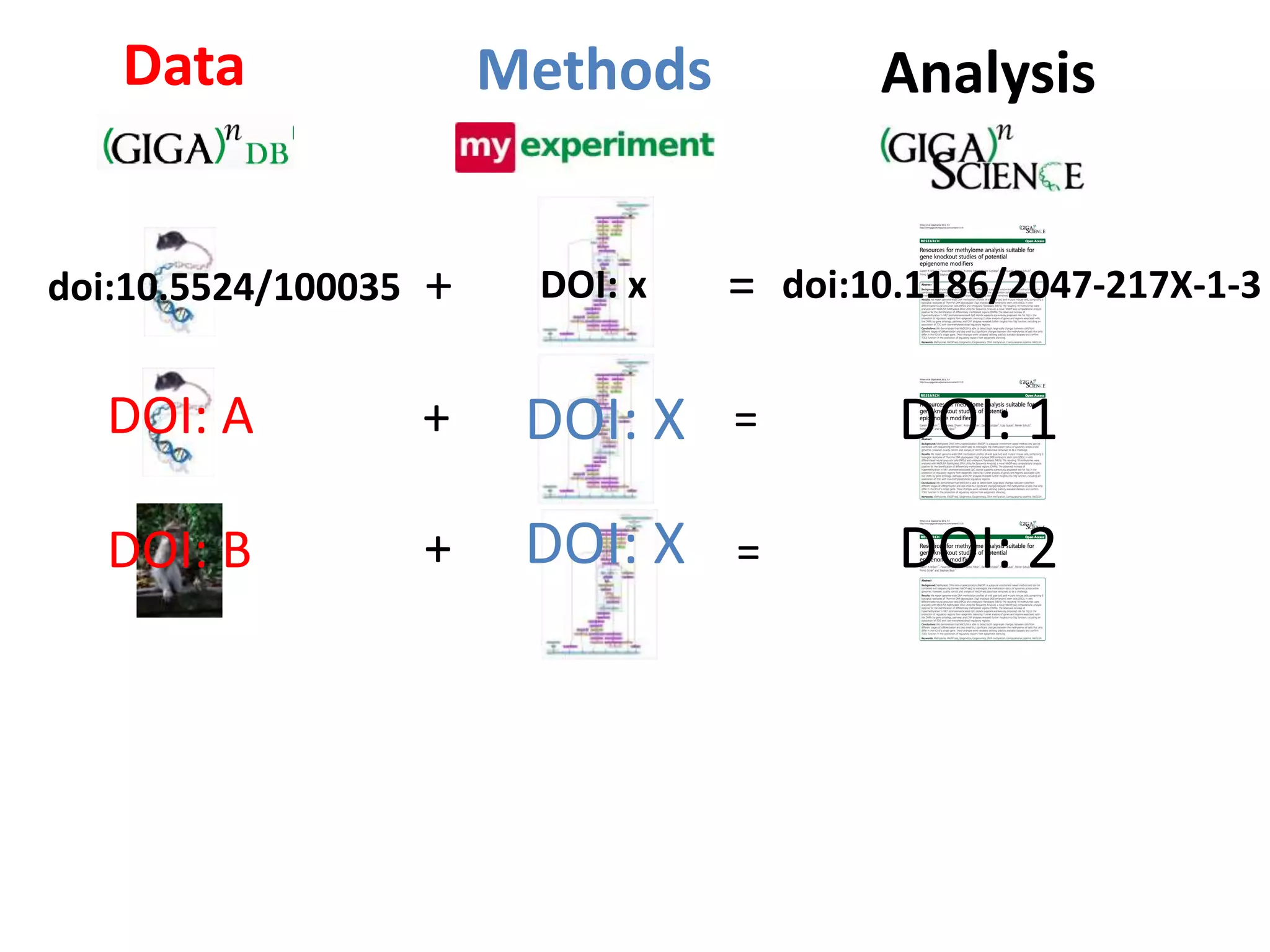
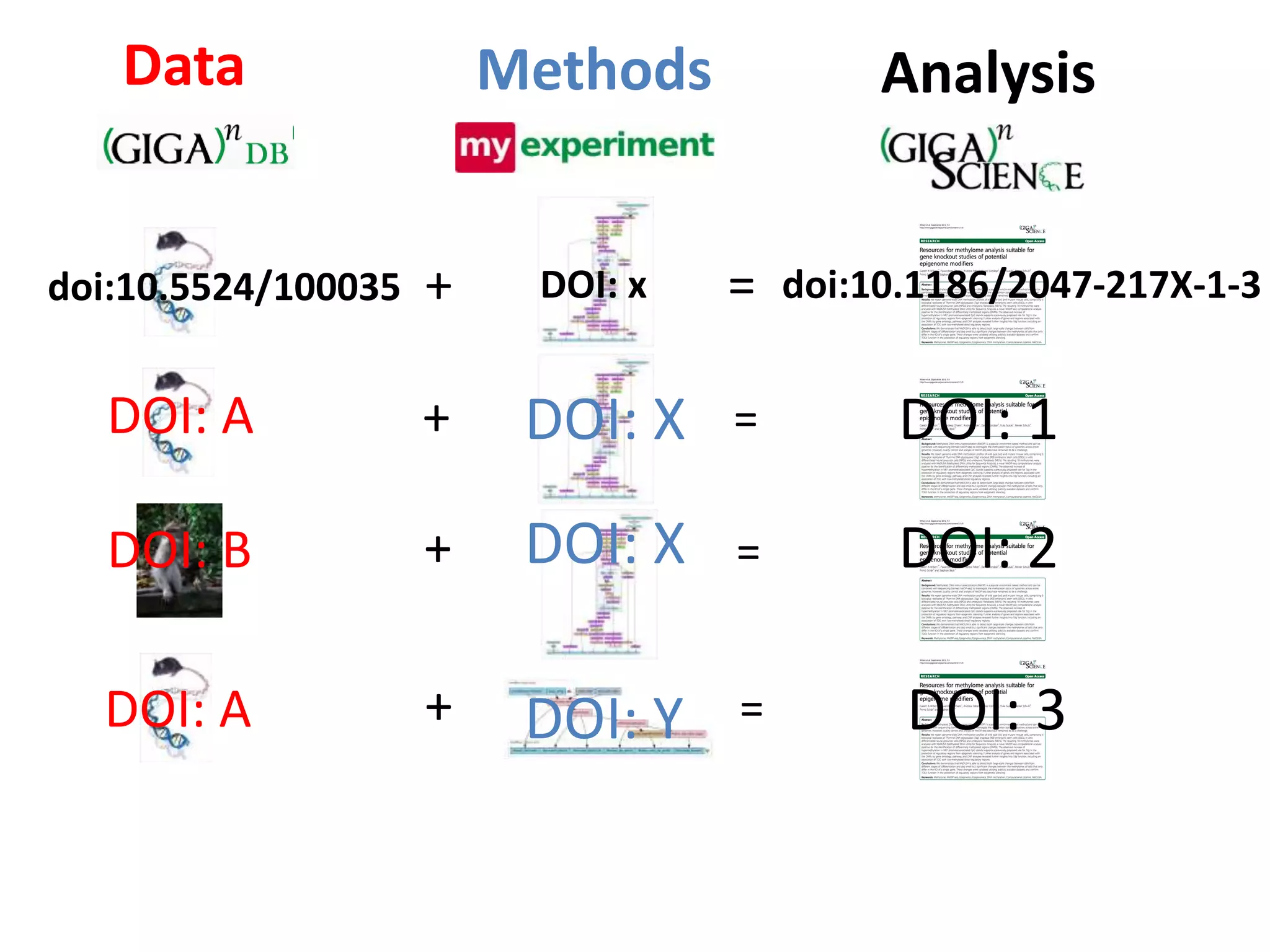
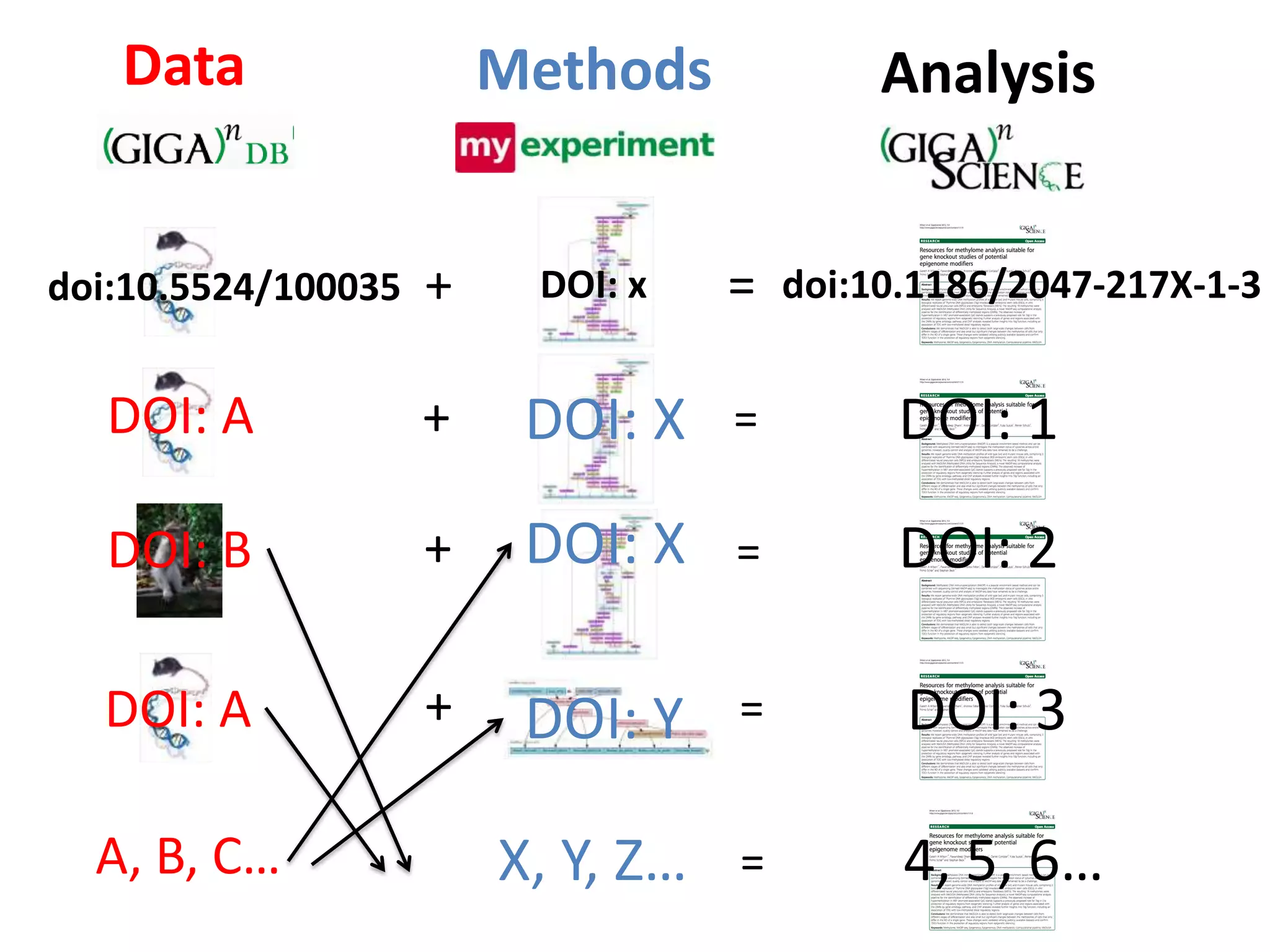
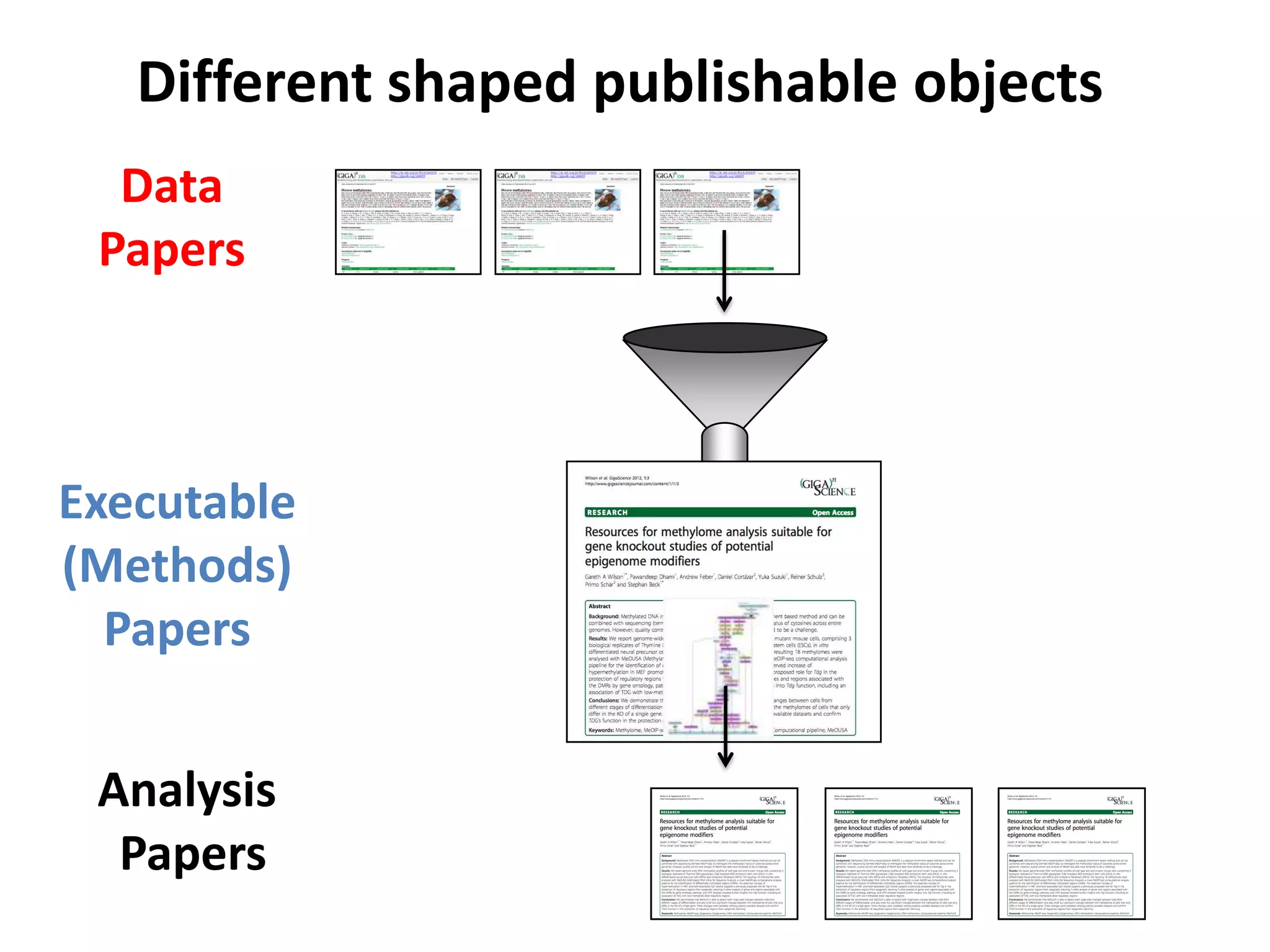
The document discusses the challenges of integrating scientific papers with data, highlighting technical issues such as data volume and cultural issues like lack of incentives for data sharing. It emphasizes the importance of transparency and reproducibility in research, especially in the context of rising retractions and claims of unrepeatable results. The text advocates for open data practices and improvements in data publishing to enhance research collaboration and scientific progress.




![Why is this important?
475, 267 (2011)
―Wide distribution of information is key to scientific progress,
yet traditionally, Chinese scientists have not systematically
released data or research findings, even after publication.―
―There have been widespread complaints from scientists
inside and outside China about this lack of transparency. ‖
―Usually incomplete and unsystematic, [what little supporting
data released] are of little value to researchers and there is
evidence that this drives down a paper's citation numbers.‖
Source: Nature 475, 267 (2011) http://www.nature.com/news/2011/110720/full/475267a.html?](https://image.slidesharecdn.com/sedmundsicg7-121201013830-phpapp02/75/Scott-Edmunds-Channeling-the-Deluge-Reproducibility-Data-Dissemination-in-the-Big-Data-Era-5-2048.jpg)