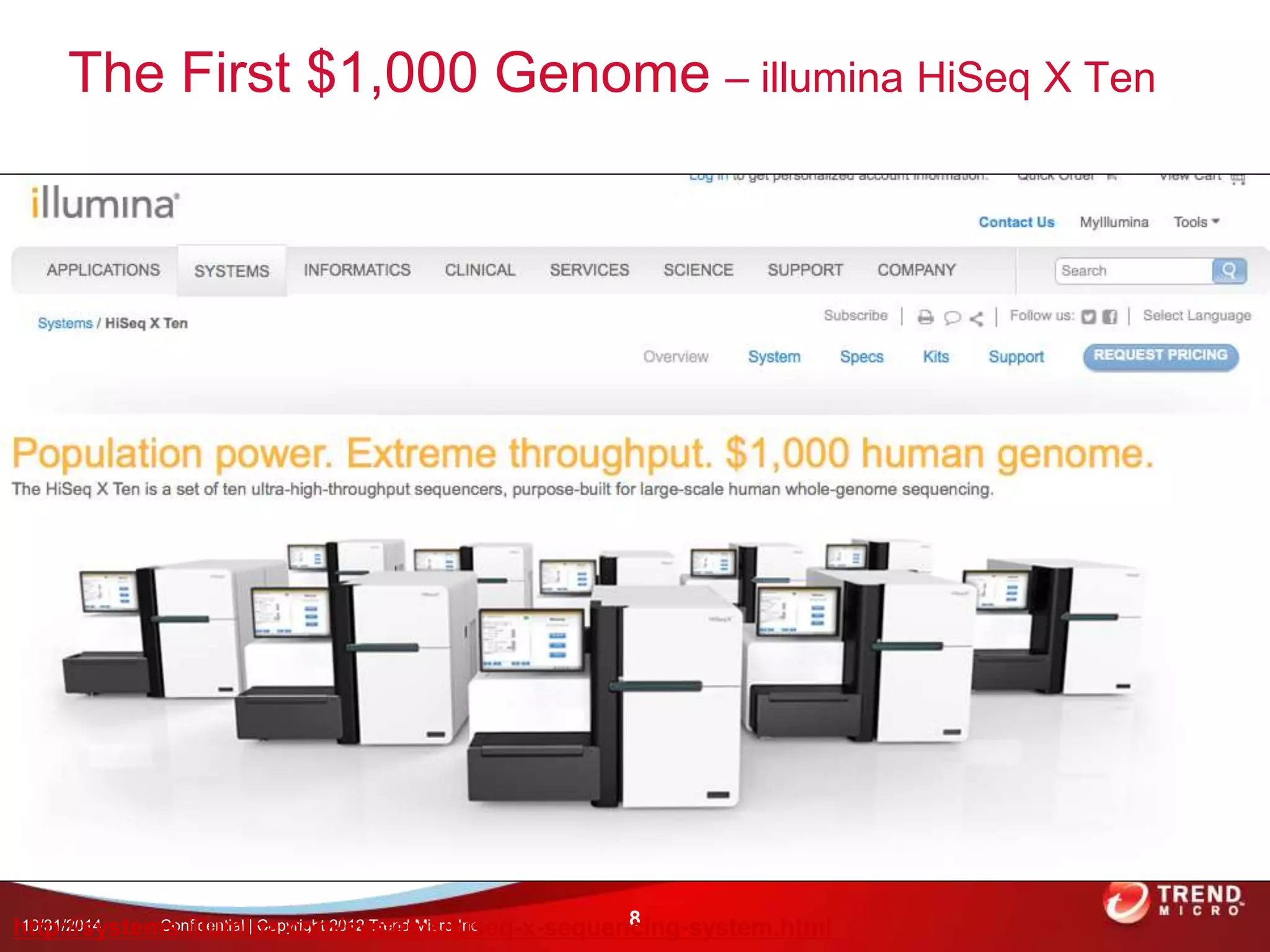
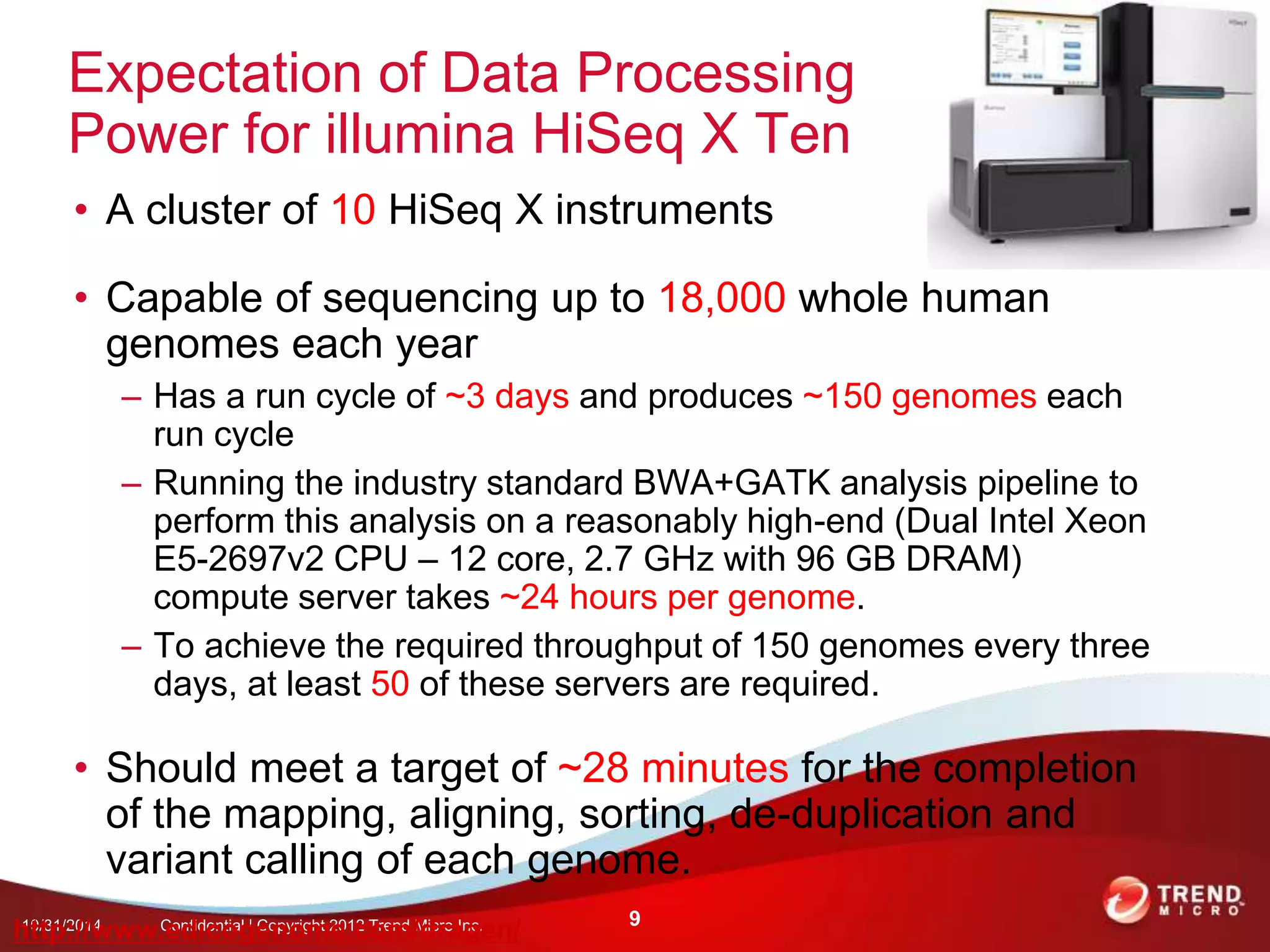
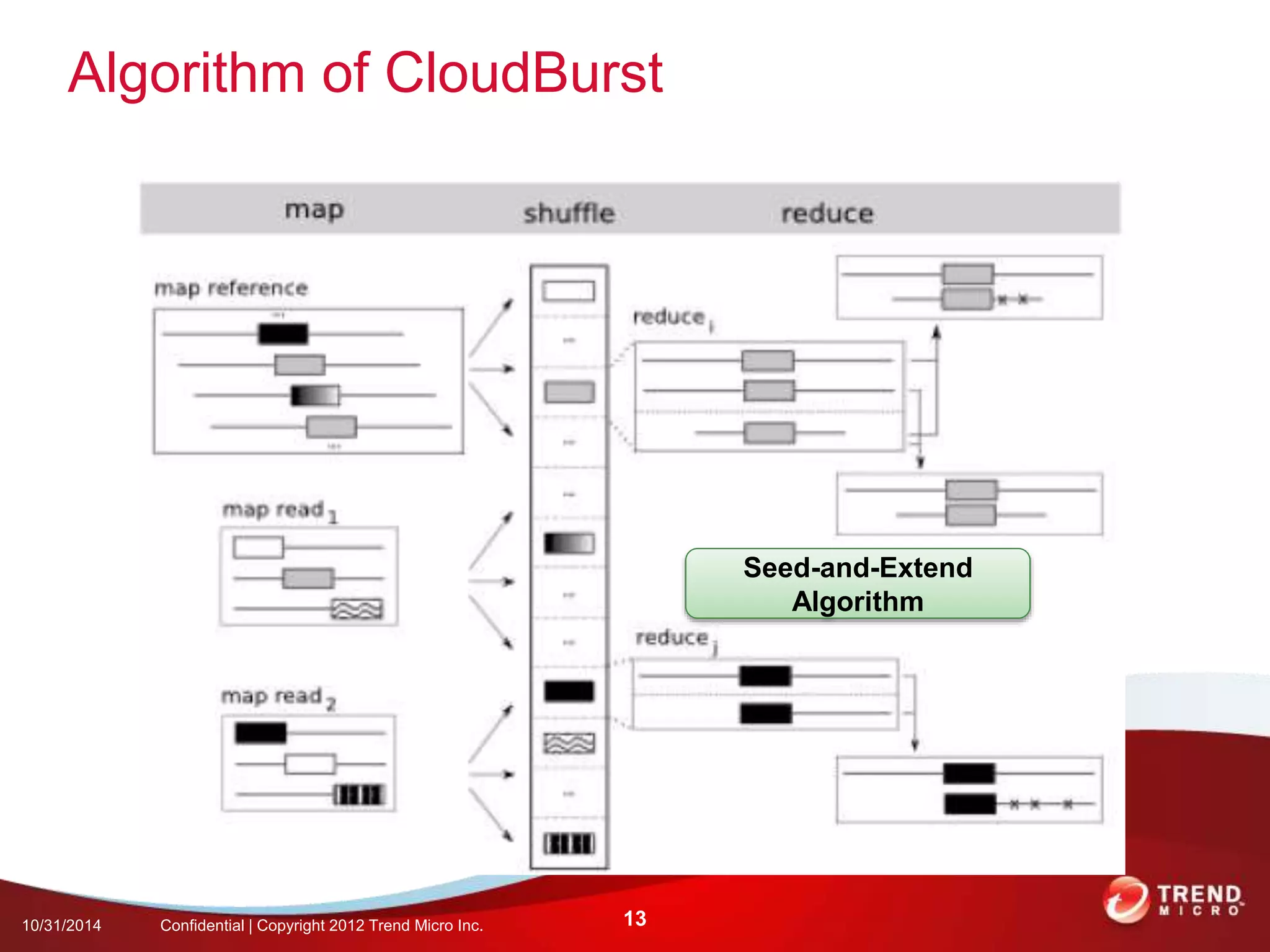
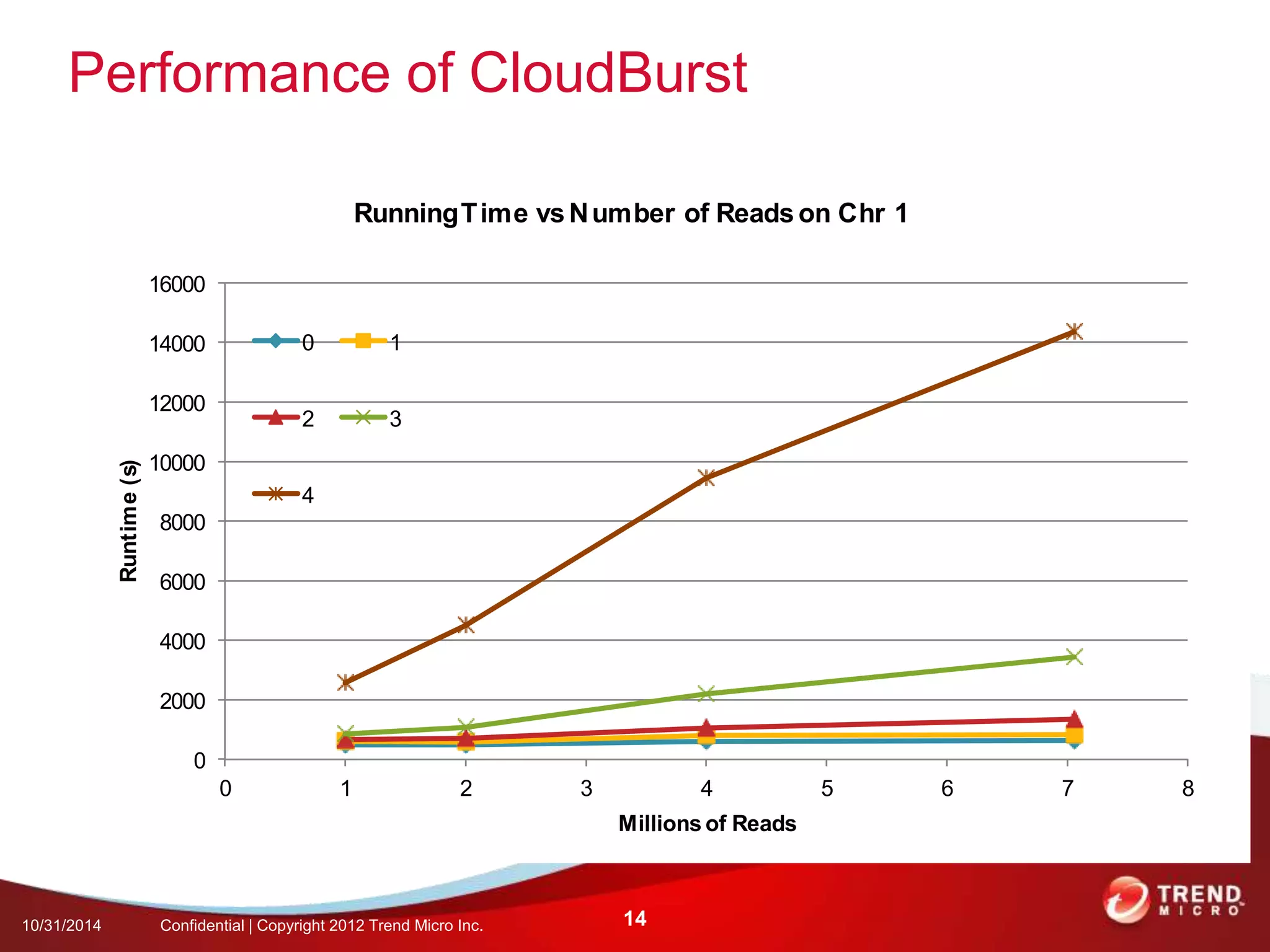
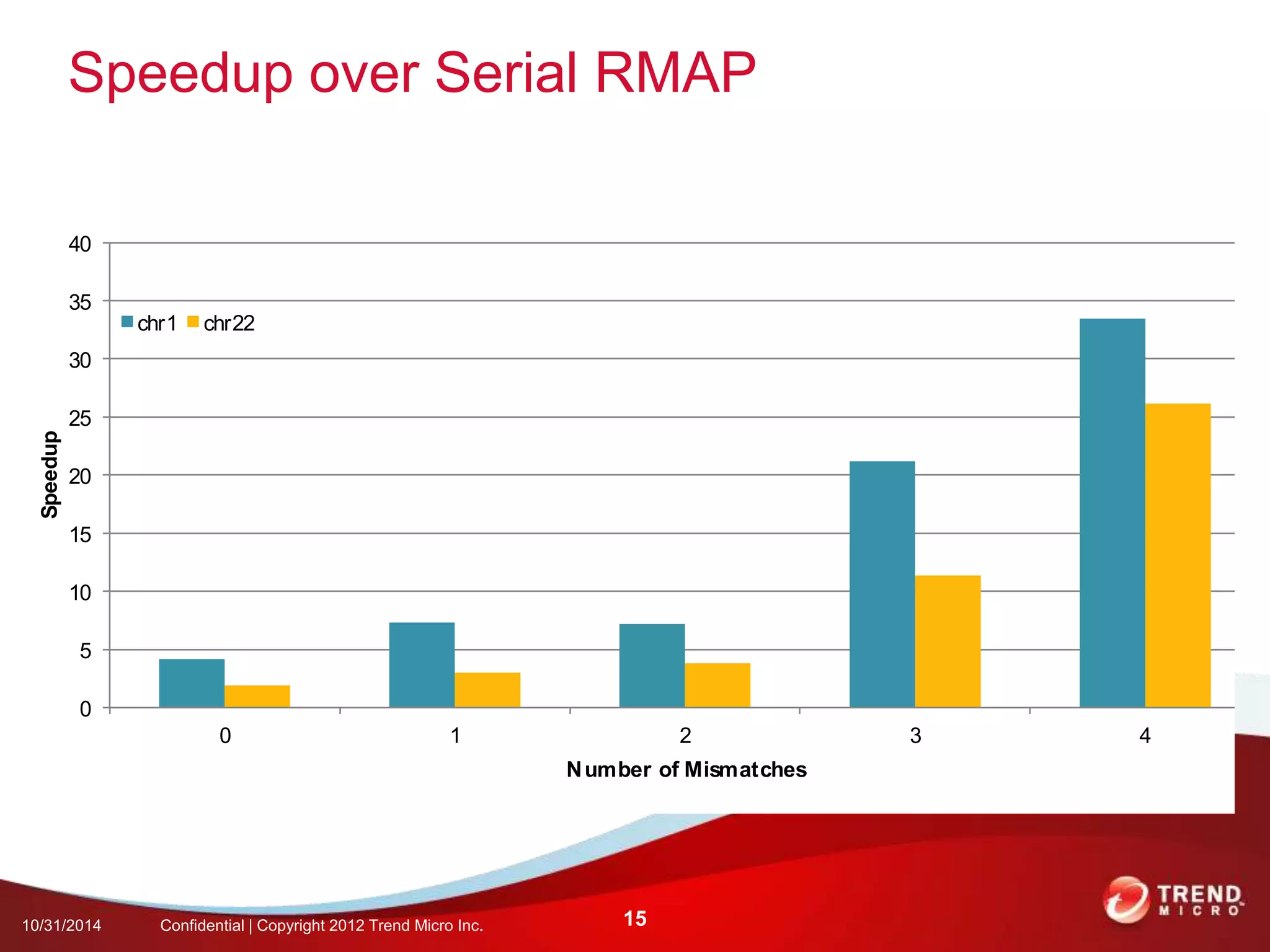
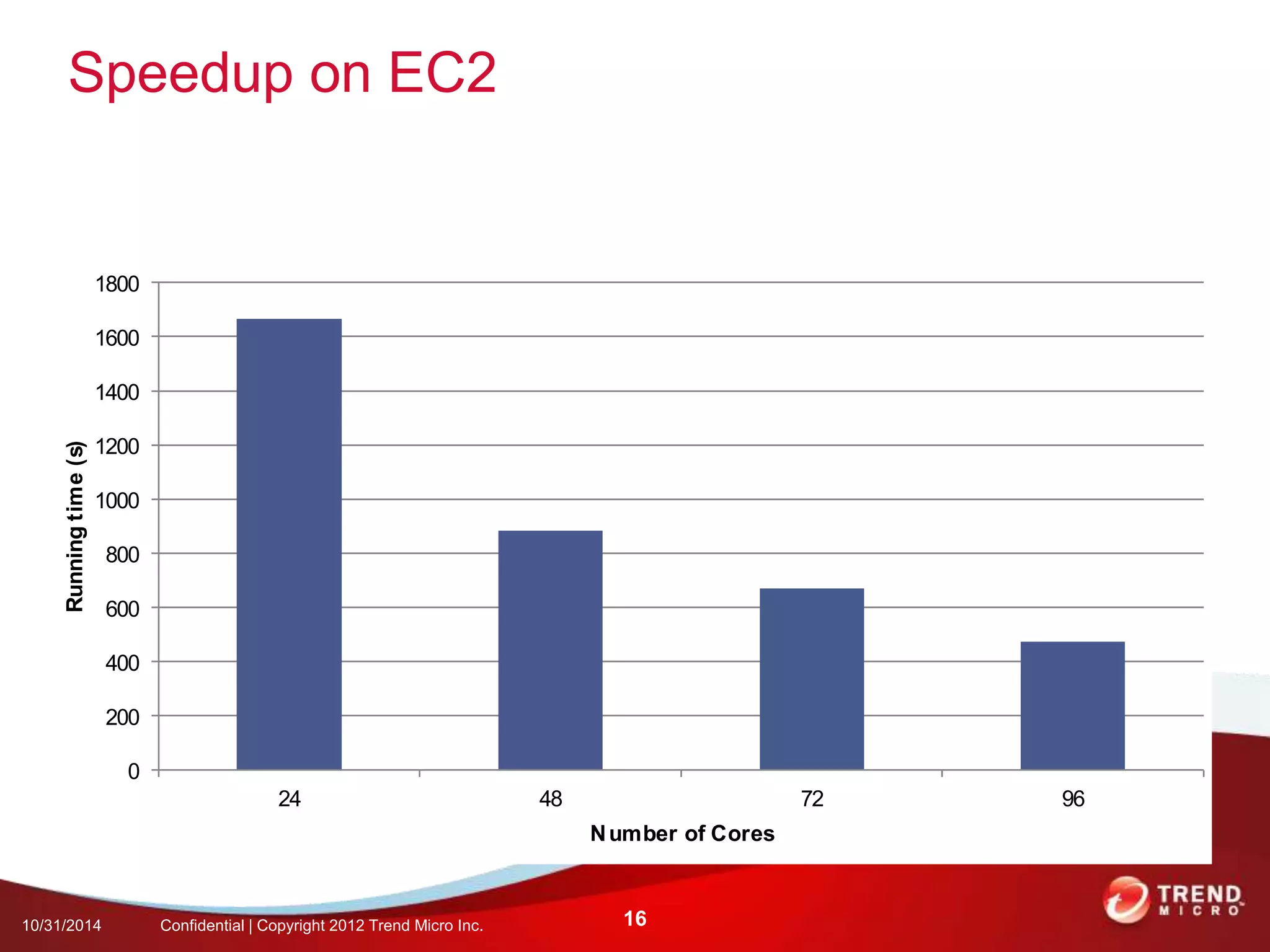
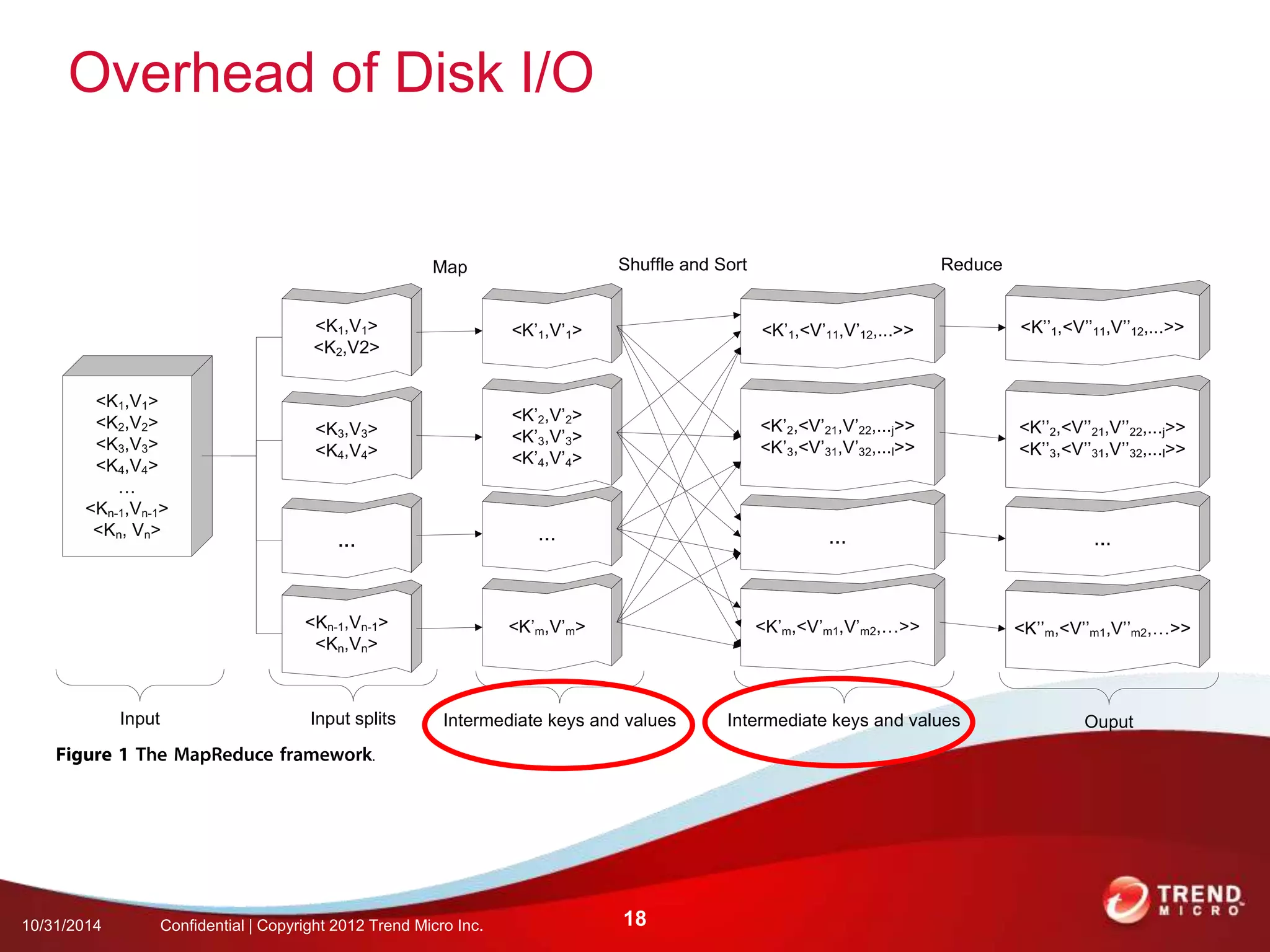
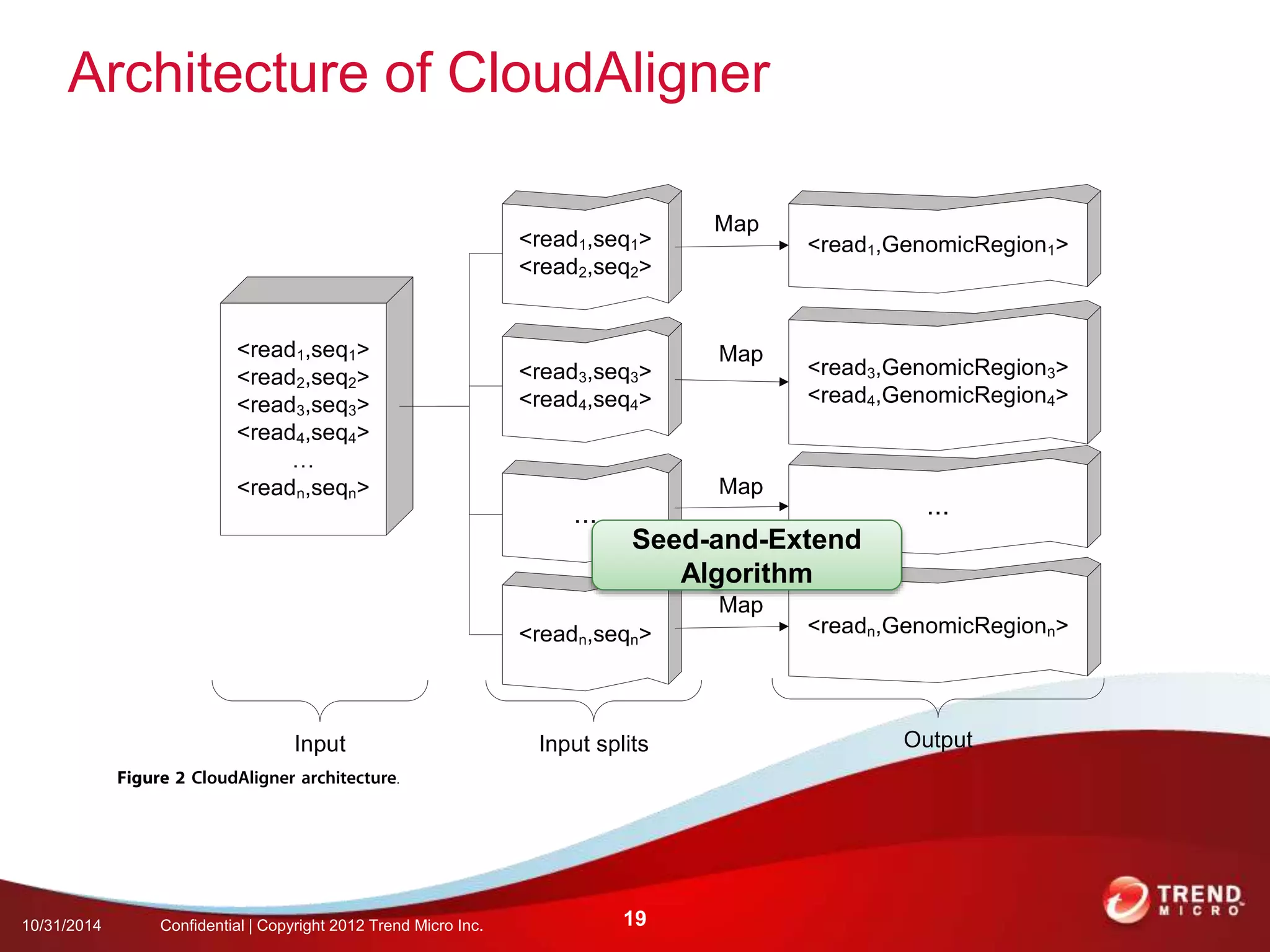
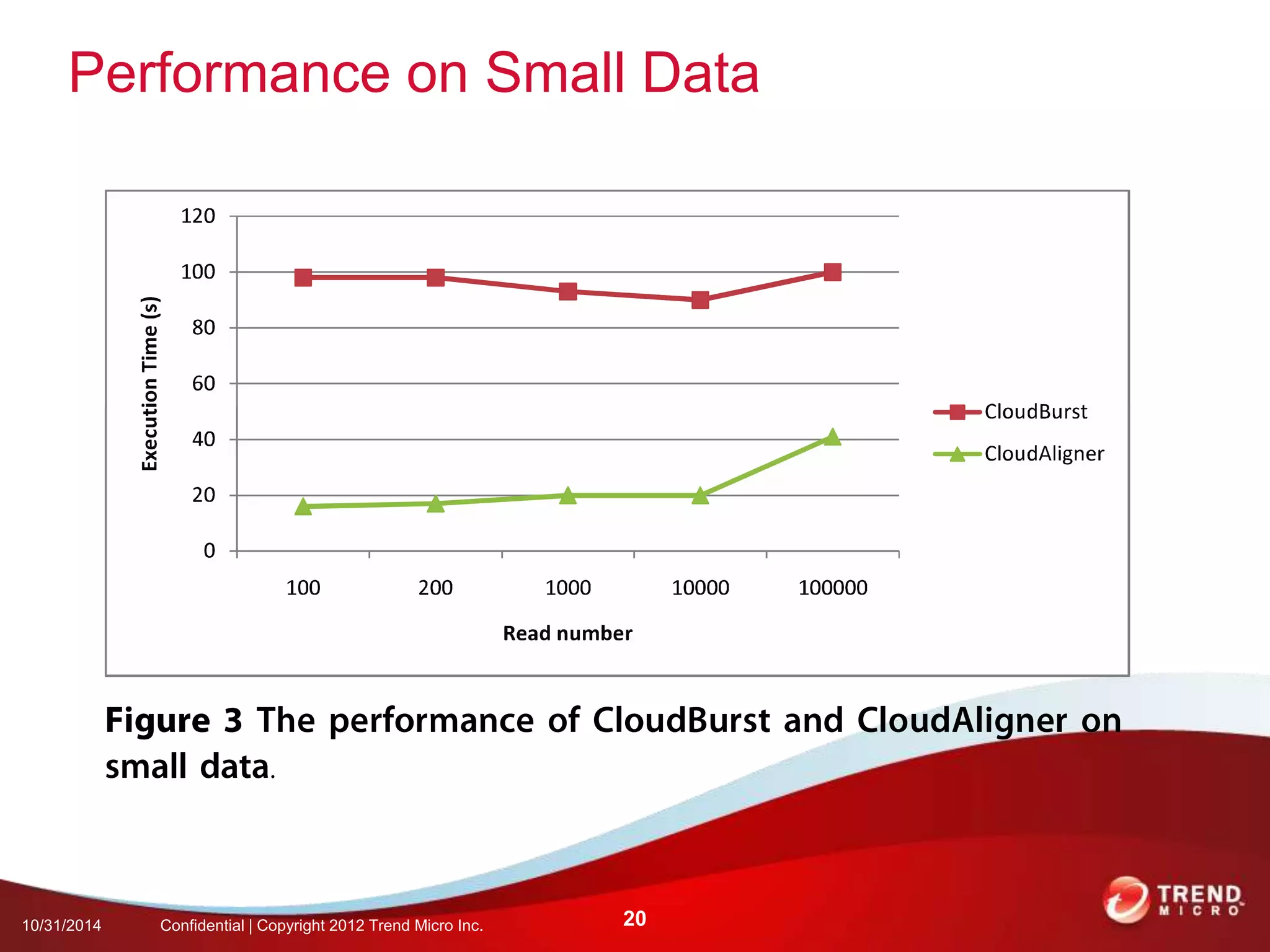
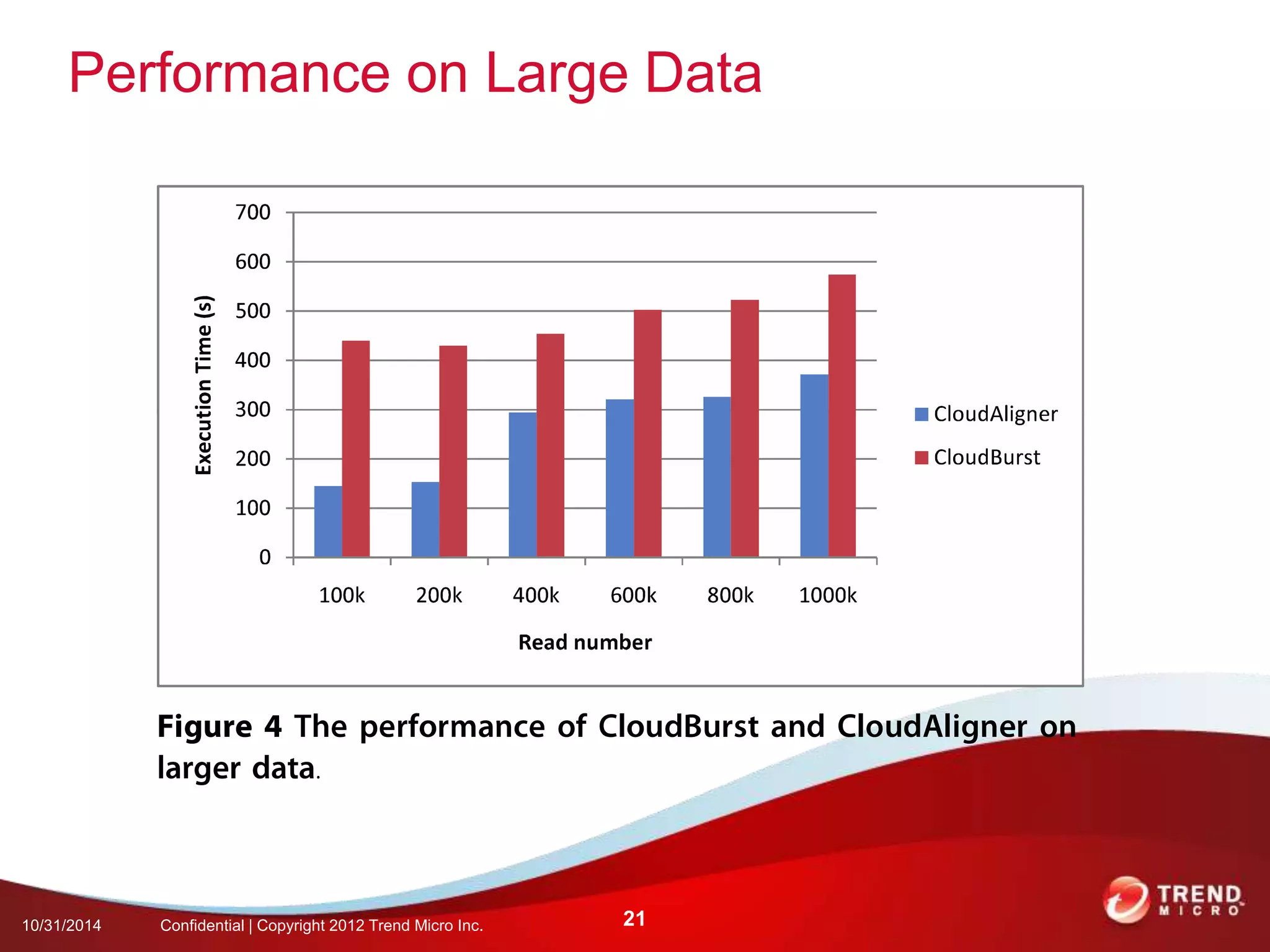
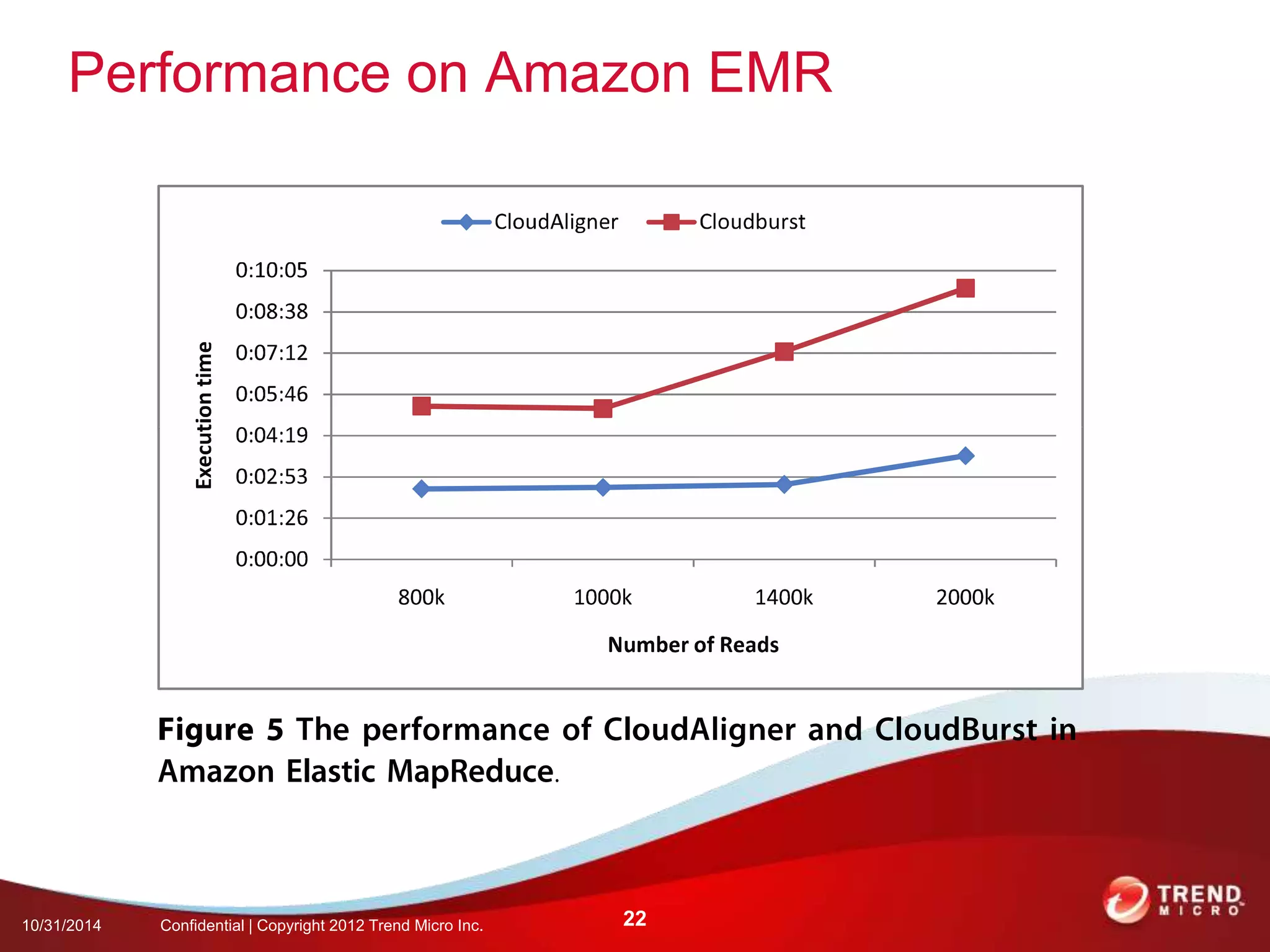
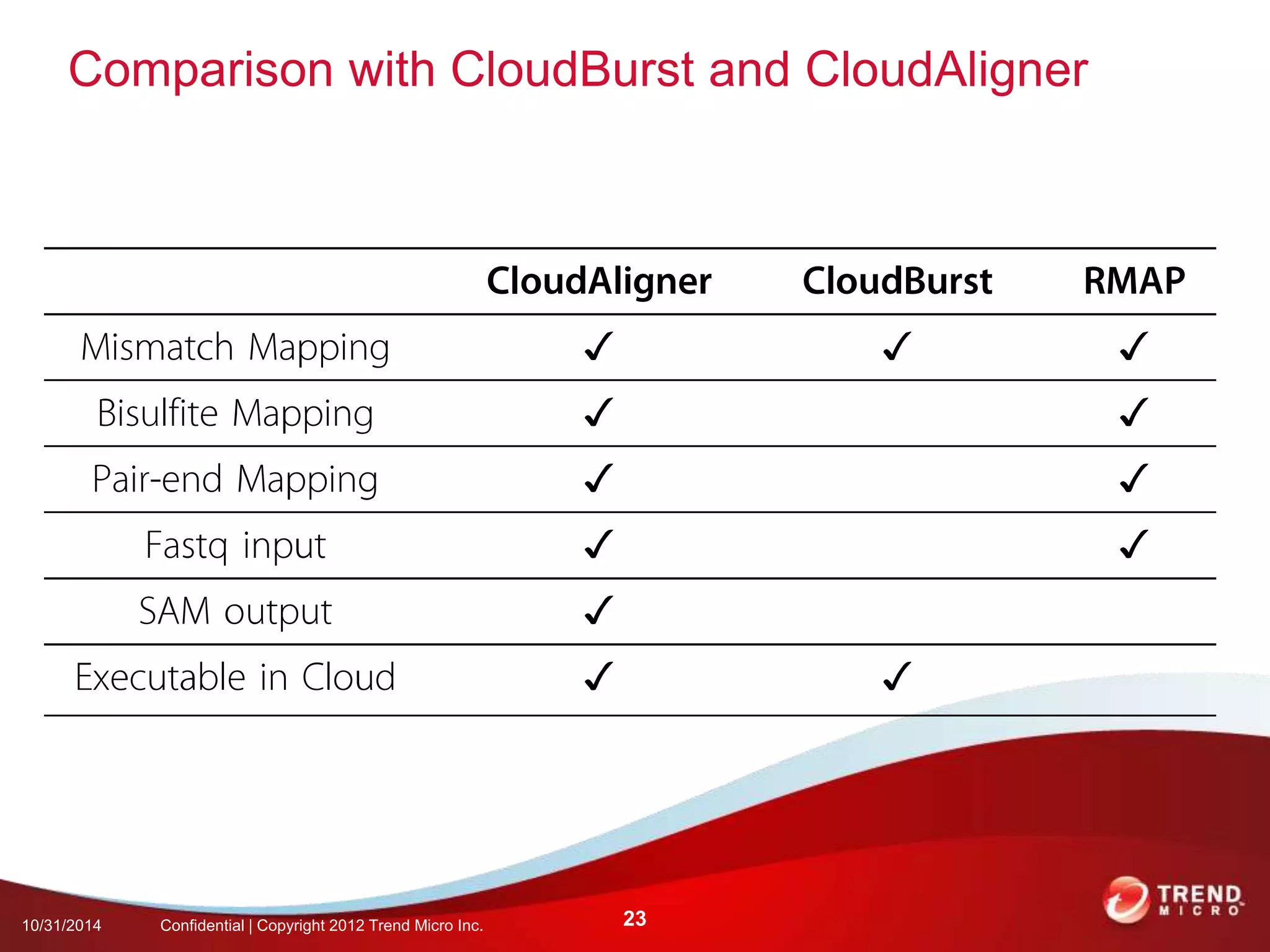
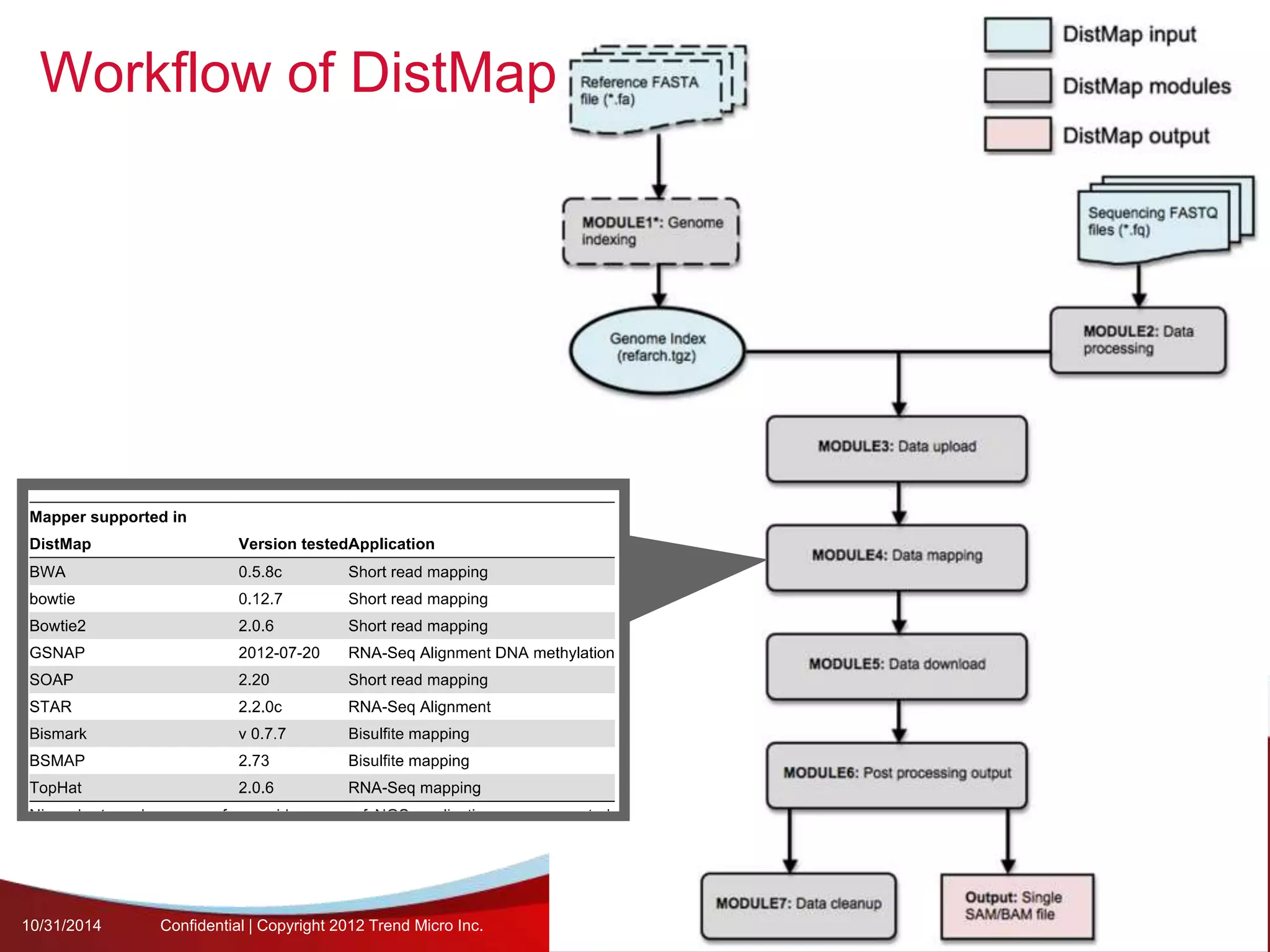
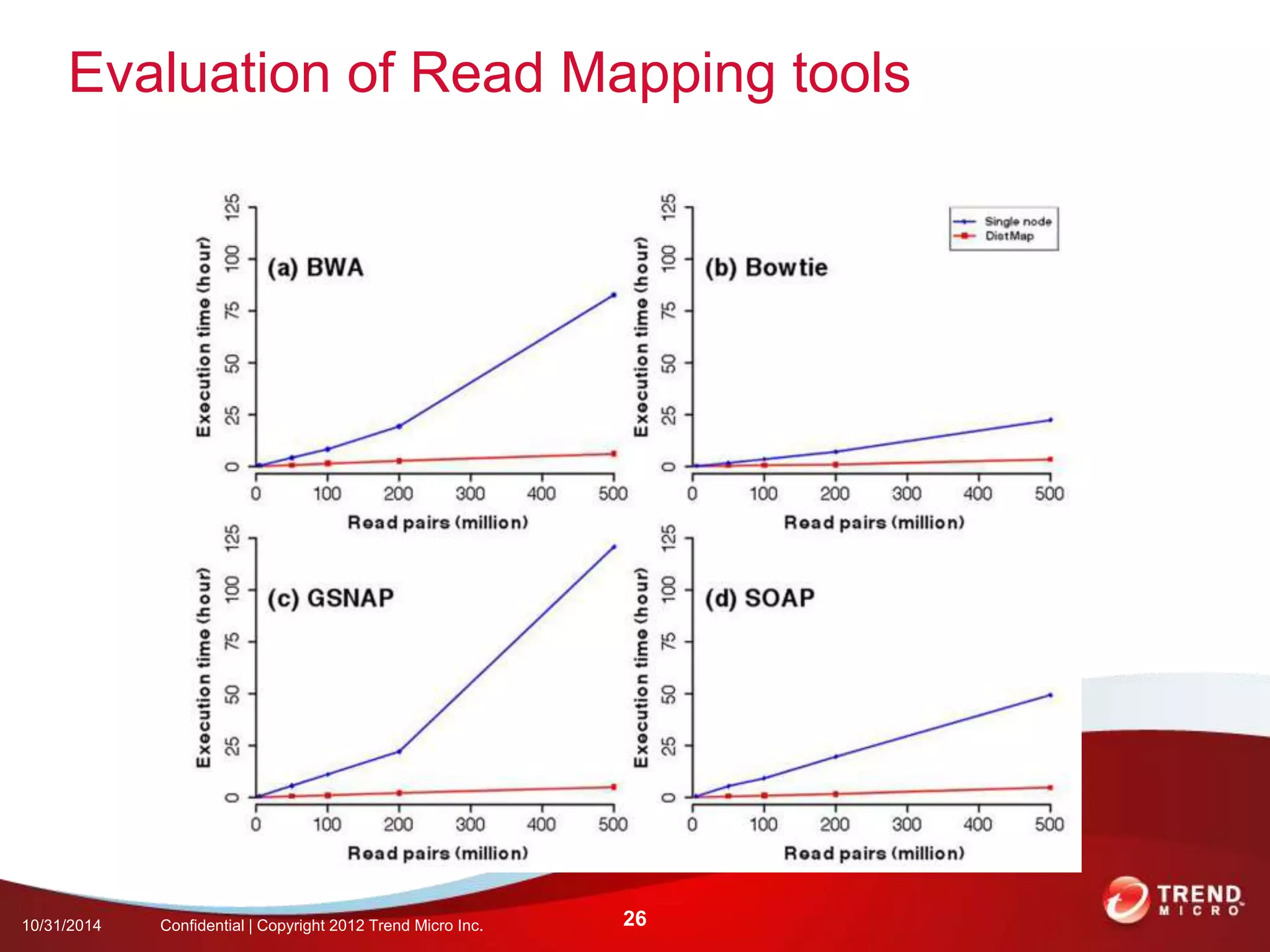
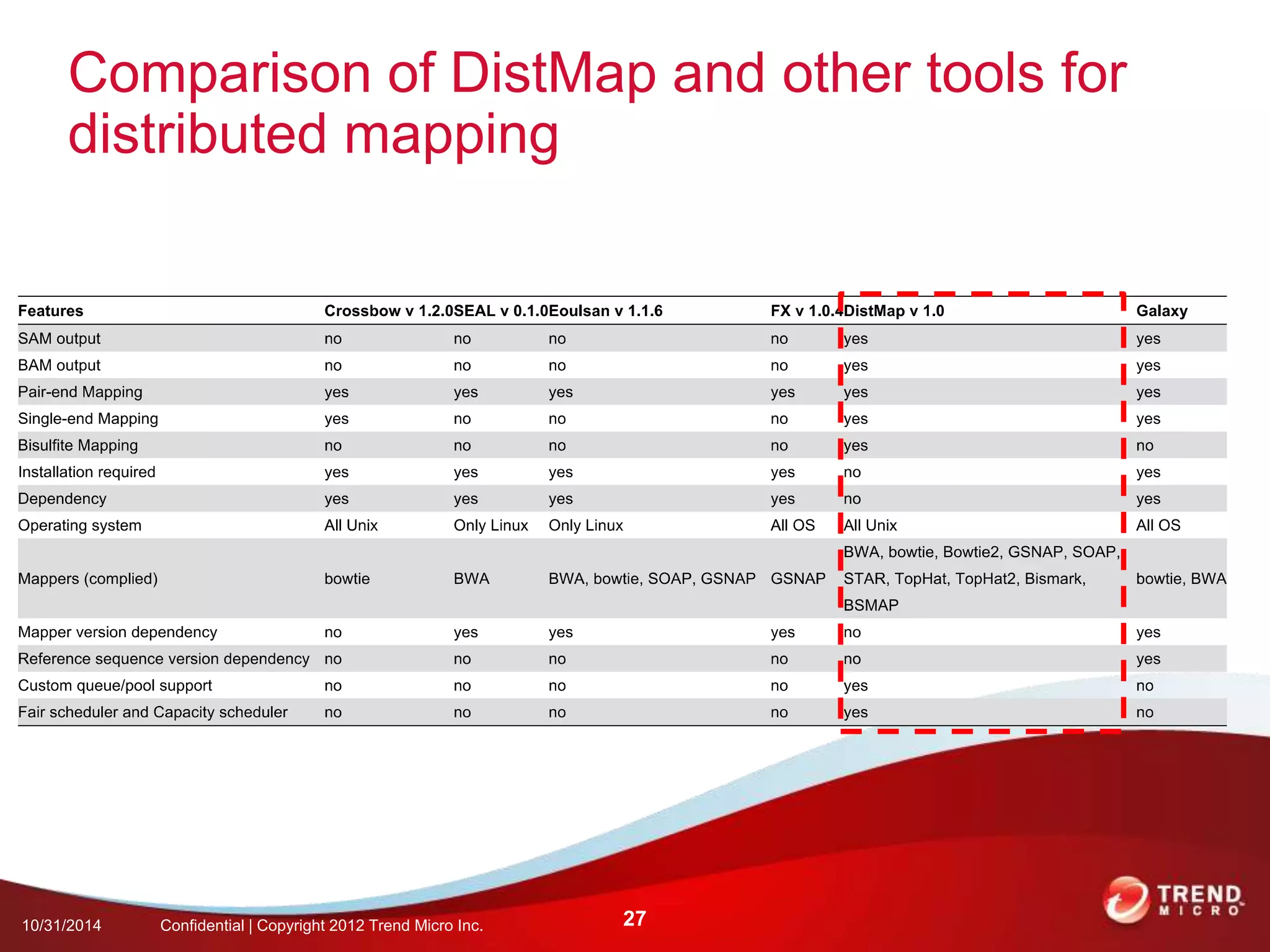
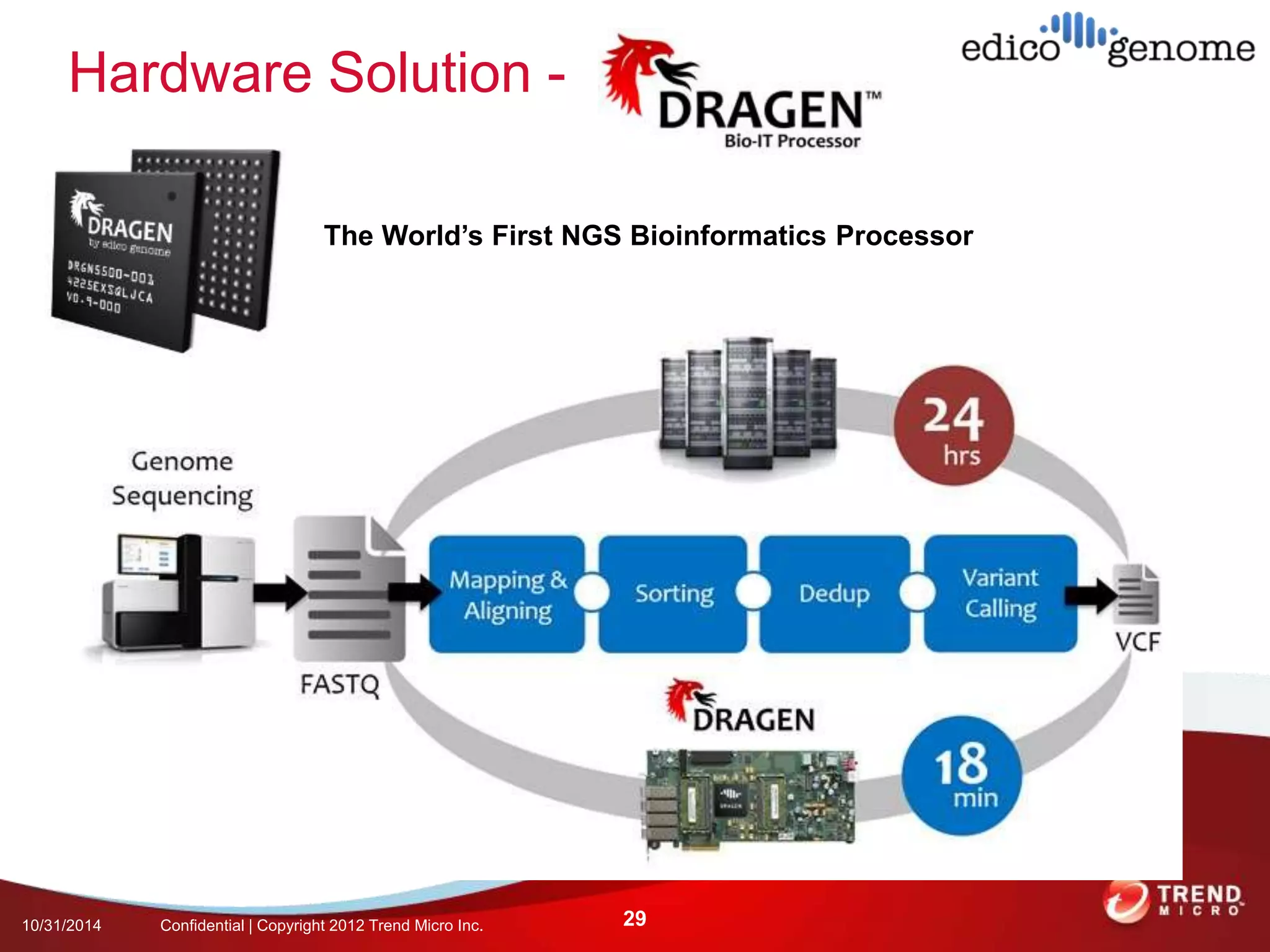
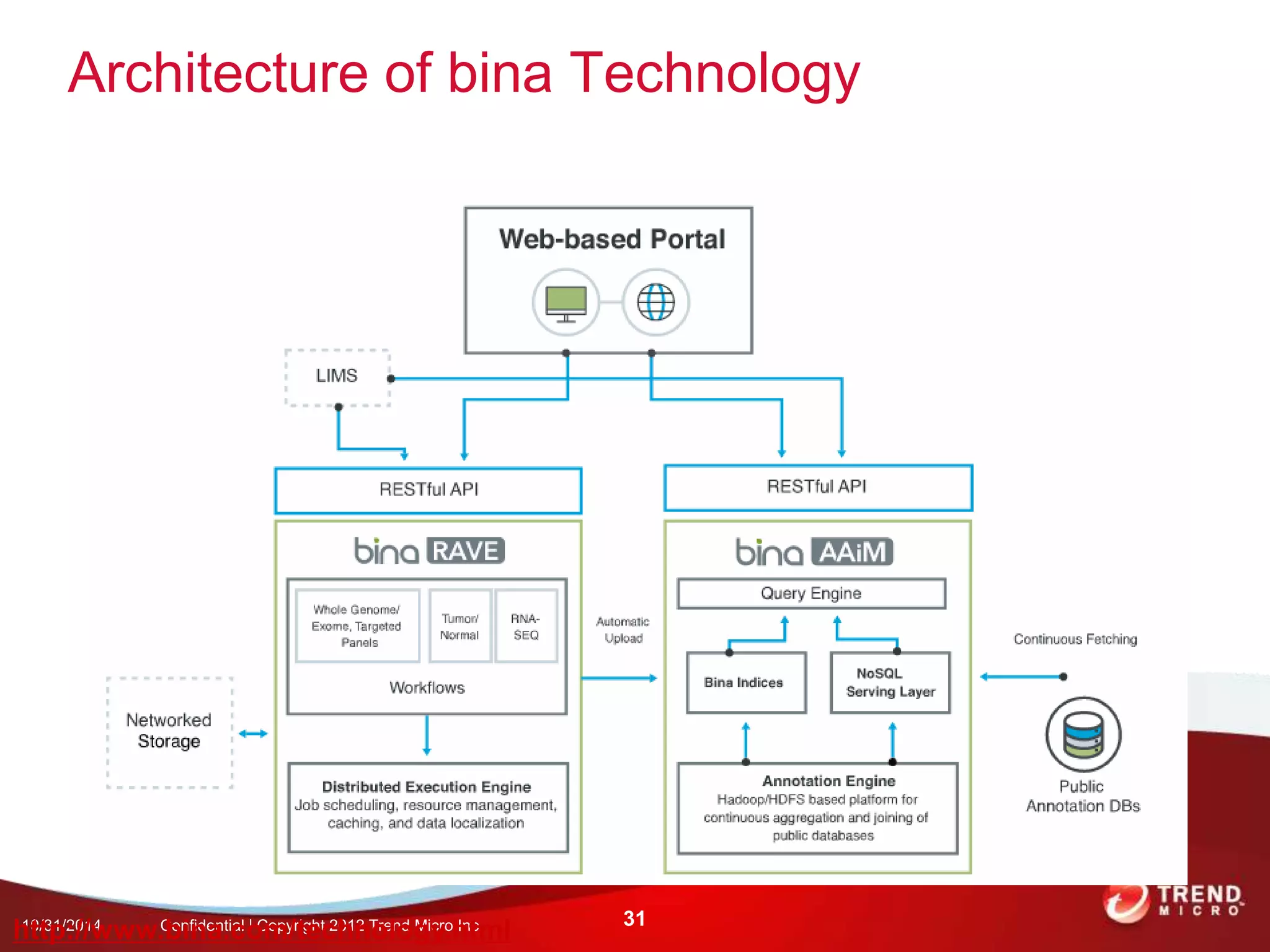
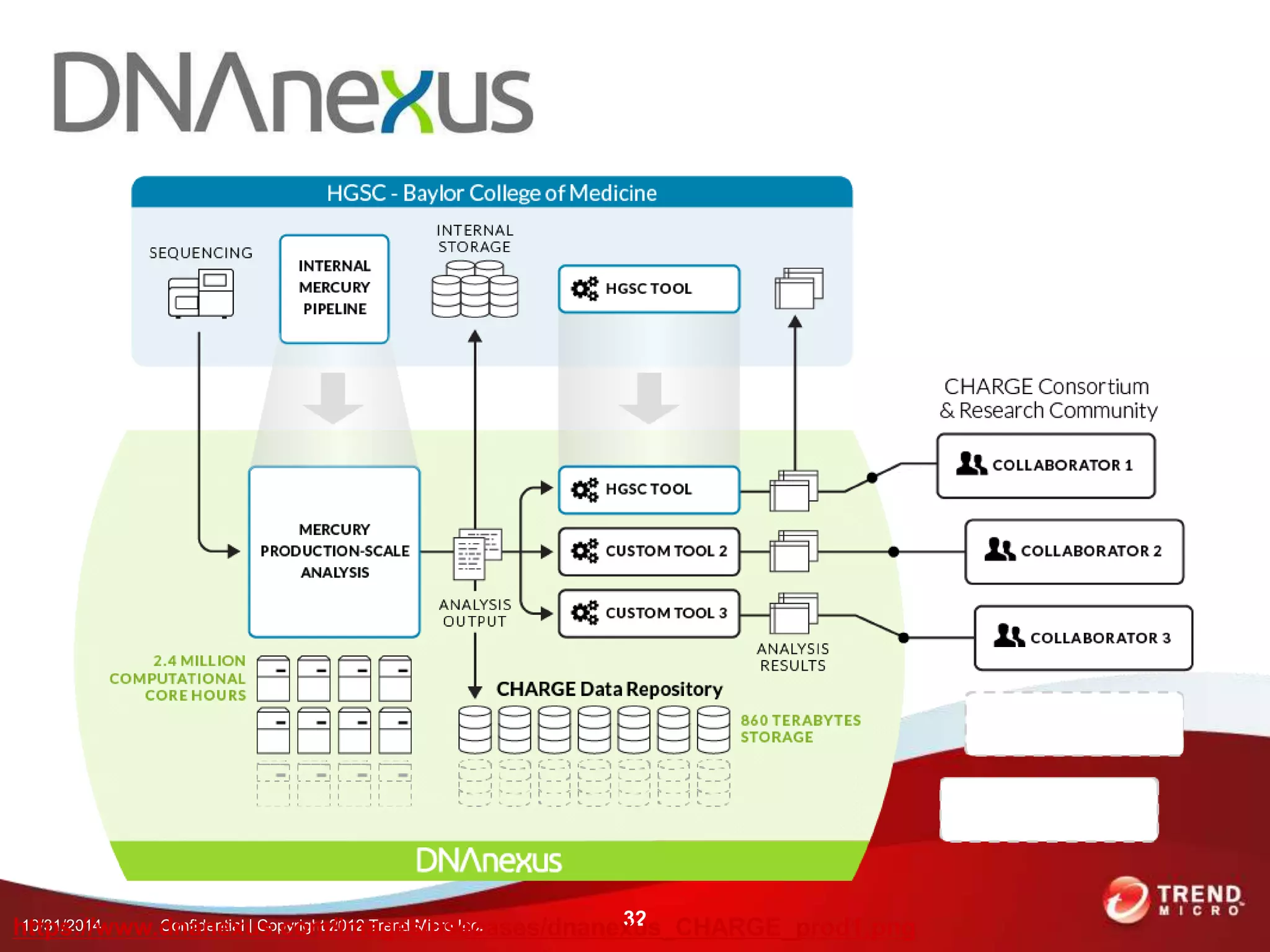
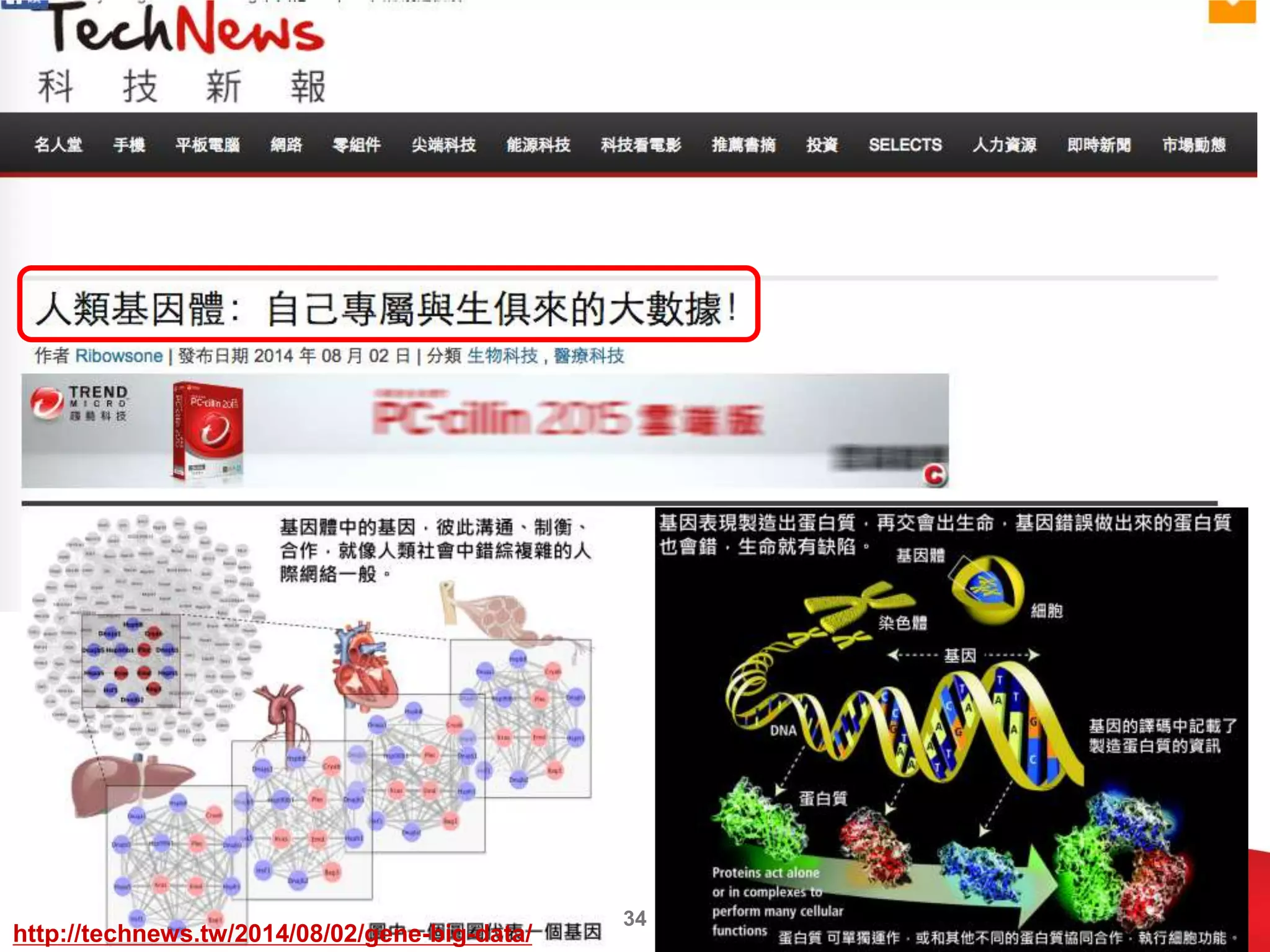
This document discusses approaches to analyzing next-generation sequencing (NGS) data on Hadoop. It summarizes three tools - CloudBurst, CloudAligner, and DistMap - that use distributed computing for mapping DNA reads to a reference genome. It also discusses challenges related to processing large NGS datasets and the need for scalable solutions. Hardware approaches like the bina processor are presented as ways to accelerate NGS analysis. Overall, the document outlines the rise of NGS as generating big genomic data and the computational challenges in analyzing this data at scale.